Temperature Effects on the Cryphonectria hypovirus 1 Accumulation and Recovery within Its Fungal Host, the Chestnut Blight Pathogen Cryphonectria parasitica
Abstract
:1. Introduction
2. Material and Methods
2.1. Viral and Fungal Strains
2.2. Culture Conditions
2.3. RNA Extraction and Reverse Transcription PCR
2.4. Statistical Analyses
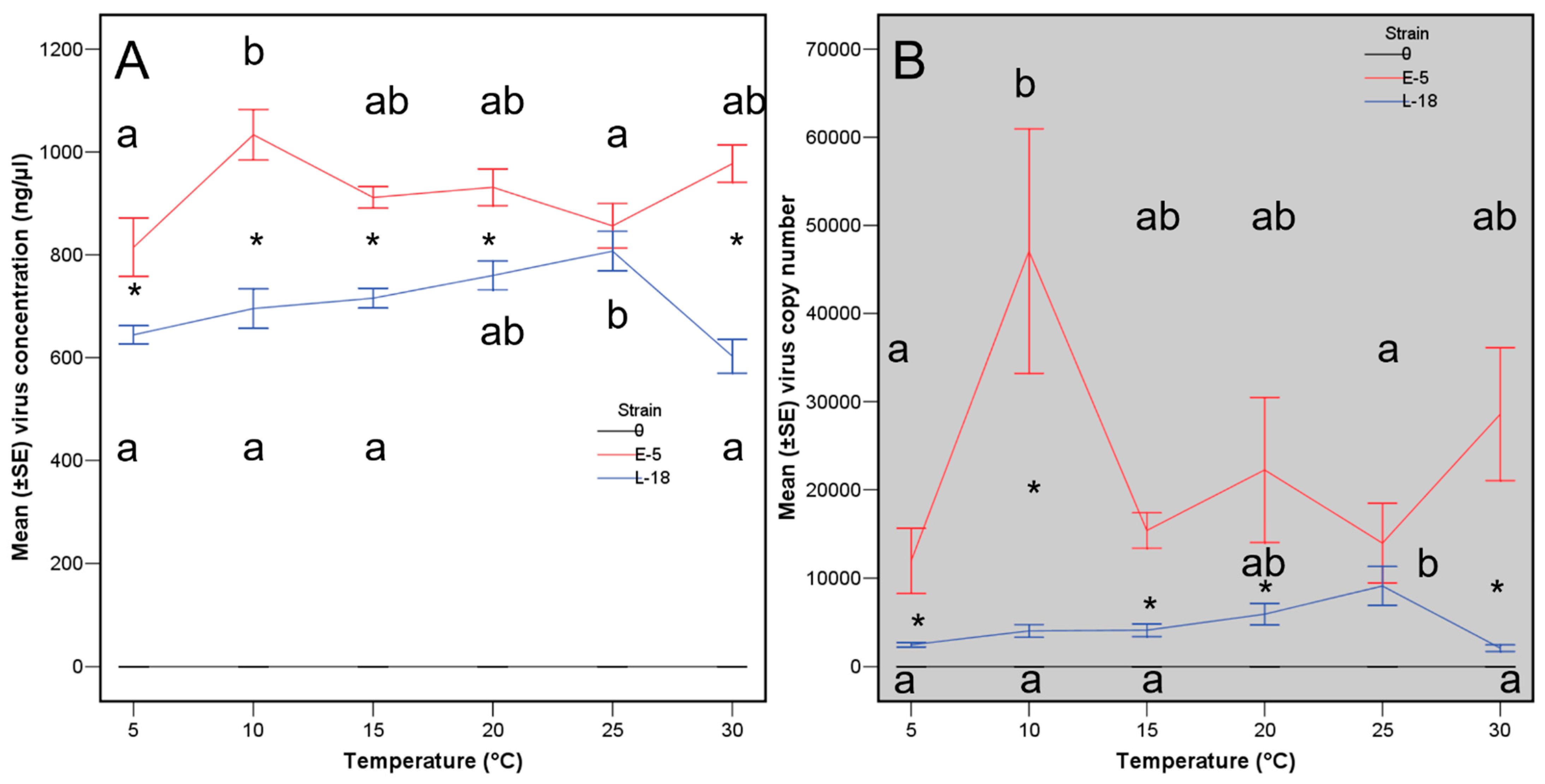
3. Results
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ghabrial, S.A.; Suzuki, N. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.N.; Beever, R.E.; Boine, B.; Arthur, K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 2009, 10, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Botella, L.; Jung, M.H.; Rost, M.; Jung, T. Natural populations from the Phytophthora palustris complex show a high diversity and abundance of ssRNA and dsRNA viruses. J. Fungi 2022, 8, 1118. [Google Scholar] [CrossRef]
- Xu, Z.; Khalifa, M.E.; Frampton, R.A.; Smith, G.R.; McDougal, R.L.; MacDiarmid, R.M.; Kalamorz, F. Characterization of a novel double-stranded RNA virus from Phytophthora pluvialis in New Zealand. Viruses 2022, 14, 247. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV). Report on Virus Classification and Taxon Nomenclature. Available online: https://ictv.global/report# (accessed on 18 May 2023).
- Hillman, B.I.; Shapira, R.; Nuss, D.L. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 1990, 80, 950–956. [Google Scholar] [CrossRef]
- Preisig, O.; Moleleki, N.; Smit, W.A.; Wingfield, B.D.; Wingfield, M.J. A novel RNA mycovirus in a hypovirulent isolate of the plant pathogen Diaporthe ambigua. J. Gen. Virol. 2000, 81, 3107–3114. [Google Scholar] [CrossRef]
- Chu, Y.; Jeon, J.; Yea, S.; Kim, Y.; Yun, S.; Lee, Y.; Kim, K. Double-stranded RNA mycovirus from Fusarium graminearum. Appl. Environ. Microbiol. 2002, 68, 2529–2534. [Google Scholar] [CrossRef]
- Chiba, S.; Salaipeth, L.; Lin, Y.H.; Sasaki, A.; Kanematsu, S.; Suzuki, N. A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: Molecular and biological characterization, taxonomic considerations, and potential for biological control. J. Virol. 2009, 83, 12801–12812. [Google Scholar] [CrossRef]
- Schoebel, C.N.; Botella, L.; Lygis, V.; Rigling, D. Population genetic analysis of a parasitic mycovirus to infer the invasion history of its fungal host. Mol. Ecol. 2017, 26, 2482–2497. [Google Scholar] [CrossRef]
- Ahn, I.P.; Lee, Y.H. A viral double-stranded RNA up regulates the fungal virulence of Nectria radicicola. Mol. Plant-Microbe Interact. 2001, 14, 496–507. [Google Scholar] [CrossRef]
- Liu, Y.C.; Dynek, J.N.; Hillman, B.I.; Milgroom, M.G. Diversity of viruses in Cryphonectria parasitica and C. nitschkei in Japan and China, and partial characterization of a new chrysovirus species. Mycol. Res. 2007, 111, 433–442. [Google Scholar] [CrossRef]
- Anagnostakis, S.L. Chestnut blight: The classical problem of an introduced pathogen. Mycologia 1987, 79, 23–37. [Google Scholar] [CrossRef]
- EPPO. Cryphonectria Parasitica. Bull. EPPO 2005, 35, 295–298. [Google Scholar] [CrossRef]
- Pérez-Sierra, A.; Romon-Ochoa, P.; Gorton, C.; Lewis, A.; Rees, H.; Van Der Linde, S.; Webber, J. High vegetative compatibility diversity of Cryphonectria parasitica infecting sweet chestnut (Castanea sativa) in Britain indicates multiple pathogen introductions. Plant Pathol. 2019, 68, 727–737. [Google Scholar] [CrossRef]
- Rigling, D.; Prospero, S. Cryphonectria parasitica, the causal agent of chestnut blight: Invasion history, population biology and disease control. Mol. Plant Pathol. 2018, 19, 7–20. [Google Scholar] [CrossRef]
- Choi, G.H.; Nuss, D.L. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 1992, 257, 800–803. [Google Scholar] [CrossRef]
- Fahima, T.; Kazmierczak, P.; Hansen, D.R.; Pfeiffer, P.; Vanalfen, N.K. Membrane-associated replication of an unencapsidated double-strand RNA of the fungus, Cryphonectria parasitica. Virology 1993, 195, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Peever, T.L.; Liu, Y.C.; Cortesi, P.; Milgroom, M.G. Variation in tolerance and virulence in the chestnut blight fungus–hypovirus interaction. Appl. Environ. Microbiol. 2000, 66, 4863–4869. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Milgroom, M.G. Correlation between hypovirus transmission and the number of vegetative incompatibility (vic) genes different among isolates from a natural population of Cryphonectria parasitica. Phytopathology 1996, 86, 79–86. [Google Scholar] [CrossRef]
- Cortesi, P.; McCulloch, C.E.; Song, H.; Lin, H.; Milgroom, M.G. Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria Parasitica. Genetics 2001, 159, 107–118. [Google Scholar] [CrossRef]
- Romon-Ochoa, P.; Foster, J.; Chitty, R.; Gorton, C.; Lewis, A.; Eacock, A.; Kupper, Q.; Rigling, D.; Pérez-Sierra, A. Canker development and biocontrol potential of CHV-1 infected English isolates of Cryphonectria parasitica is dependent on the virus concentration and the compatibility of the fungal inoculums. Viruses 2022, 14, 2678. [Google Scholar] [CrossRef]
- Aulia, A.; Andika, I.B.; Kondo, H.; Hillman, B.I.; Suzuki, N. A symptomless hypovirus, CHV4, facilitates stable infection of the chestnut blight fungus by a coinfecting reovirus likely through suppression of antiviral RNA silencing. Virology 2019, 533, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Romon-Ochoa, P.; Kranjec Orlovic, J.; Gorton, C.; Lewis, A.; van der Linde, S.; Pérez-Sierra, A. New detections of chestnut blight in Great Britain during 2019–2020 reveal high Cryphonectria parasitica diversity and limited spread of the disease. Plant Pathol. 2021, 71, 793–804. [Google Scholar] [CrossRef]
- Rigling, D.; Borst, N.; Cornejo, C.; Supatashvili, A.; Prospero, S. Genetic and phenotypic characterization of Cryphonectria hypovirus 1 from Eurasian Georgia. Viruses 2018, 10, 687. [Google Scholar] [CrossRef]
- Allemann, C.; Hoegger, P.; Heiniger, U.; Rigling, D. Genetic variation of Cryphonectria hypoviruses (CHV1) in Europe, assessed using restriction fragment length polymorphism (RFLP) markers. Mol. Ecol. 1999, 8, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Bryner, S.F.; Rigling, D. Hypovirus virulence and vegetative incompatibility in populations of the chestnut blight fungus. Phytopathology 2012, 102, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Krstin, L.; Novak-Agbaba, S.; Rigling, D.; Krajacic, M.; Curkovic-Perica, M. Chestnut blight fungus in Croatia: Diversity of vegetative compatibility types and genetic variability of associated Cryphonectria hypovirus 1. Plant Pathol. 2008, 57, 1086–1096. [Google Scholar] [CrossRef]
- Krstin, L.; Novak-Agbaba, S.; Rigling, D.; Curkovic-Perica, M. Diversity of vegetative compatibility types and mating types of Cryphonectria parasitica in Slovenia and occurrence of associated Cryphonectria hypovirus 1. Plant Pathol. 2011, 60, 752–761. [Google Scholar] [CrossRef]
- Sotiroski, K.; Milgroom, M.G.; Rigling, D.; Heiniger, U. Occurrence of Cryphonectria hypovirus 1 in the chestnut blight fungus in Macedonia. For. Pathol. 2006, 36, 136–143. [Google Scholar] [CrossRef]
- Akilli, S.; Serce, C.U.; Katircioglu, Y.Z.; Maden, S.; Rigling, D. Characterization of hypovirulent isolates of the chestnut blight fungus, Cryphonectria parasitica from the Marmara and Black Sea regions of Turkey. Eur. J. Plant Pathol. 2013, 135, 323–334. [Google Scholar] [CrossRef]
- Hoegger, P.J.; Heiniger, U.; Holdenrieder, O.; Rigling, D. Differential transfer and dissemination of hypovirus and nuclear and mitochondrial genomes of a hypovirus-infected Cryphonectria parasitica strain after introduction into a natural population. Appl. Environ. Microbiol. 2003, 69, 3767–3771. [Google Scholar] [CrossRef] [PubMed]
- Statista. Monthly Mean Air Temperature in England from 2015 to 2022; UK Department for Business, Energy, and Industrial Strategy: London, UK, 2022.
- Ježić, M.; Schwarz, J.M.; Prospero, S.; Sotirovski, K.; Risteski, M.; Ćurković-Perica, M.; Nuskern, L.; Krstin, L.; Katanić, Z.; Maleničić, E.; et al. Temporal and spatial genetic population structure of Cryphonectria parasitica and its associated hypovirus across an invasive range of chestnut blight in Europe. Phytopathology 2021, 111, 1327–1337. [Google Scholar] [CrossRef]
- Bryner, S.F.; Rigling, D. Temperature-dependent genotype-by-genotype interaction between a pathogenic fungus and its hyperparasitic virus. Am. Nat. 2011, 177, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Marquez, L.M.; Redman, R.S.; Rodriguez, R.; Stout, R.G.; Rodriguez, R.J.; Roossinck, M. A virus in a fungus in a plant: Three-way symbiosis required for thermal tolerance. Science 2007, 315, 513–515. [Google Scholar] [CrossRef] [PubMed]
- Vainio, E.J.; Hyder, R.; Aday, G.; Hansen, E.; Piri, T.; Doğmuş-Lehtijärvi, T.; Hantula, J. Population structure of a novel putative mycovirus infecting the conifer root-rot fungus Heterobasidion annosum sensu lato. Virology 2012, 422, 366–376. [Google Scholar] [CrossRef]
- Romon-Ochoa, P.; Lewis, A.; Gorton, C.; van der Linde, S.; Pérez-Sierra, A. Effects of growth medium, temperature and mycelium age on CHV-1 accumulation and transmission. For. Ecol. Manag. 2023, 529, 120705–120717. [Google Scholar] [CrossRef]
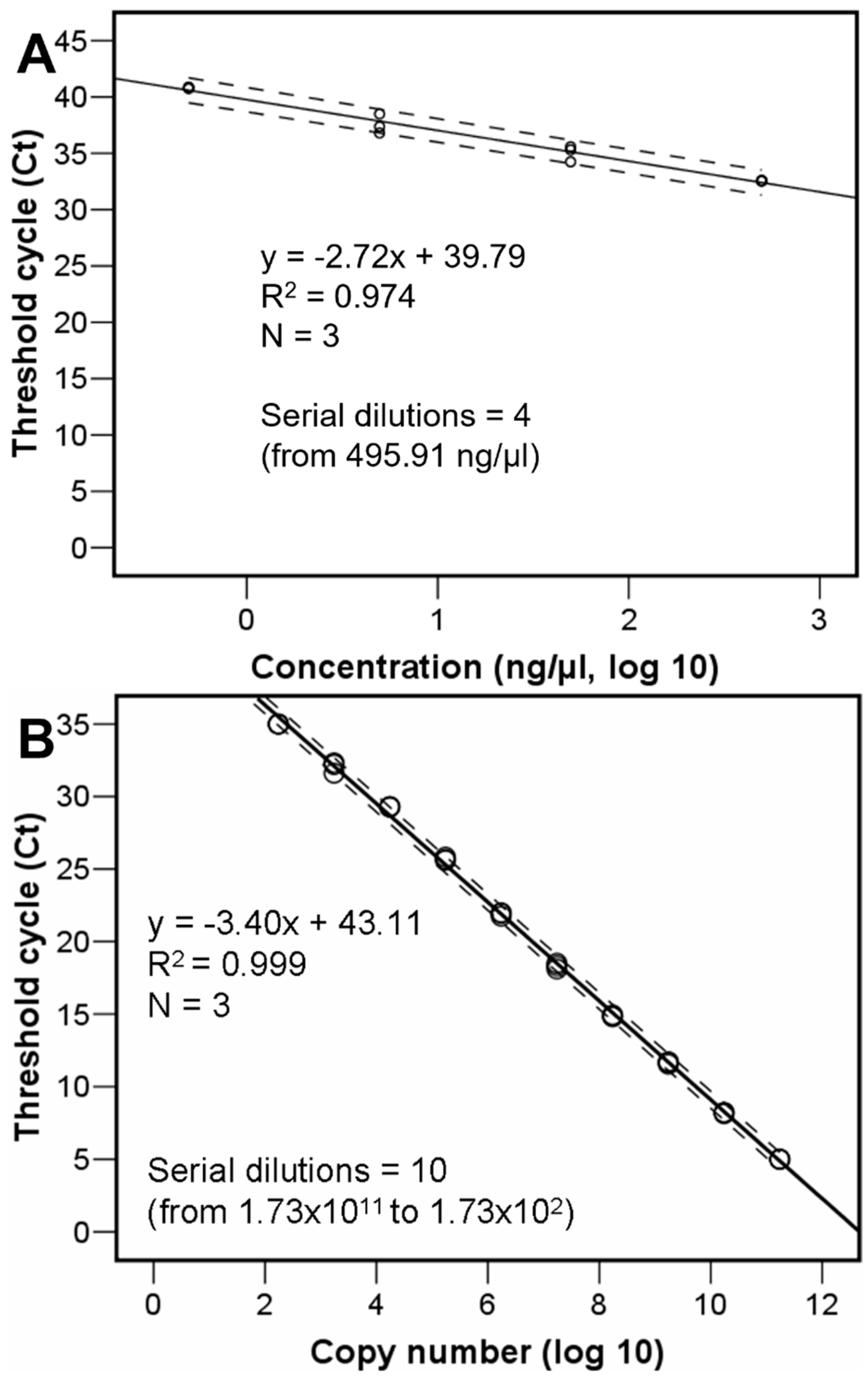

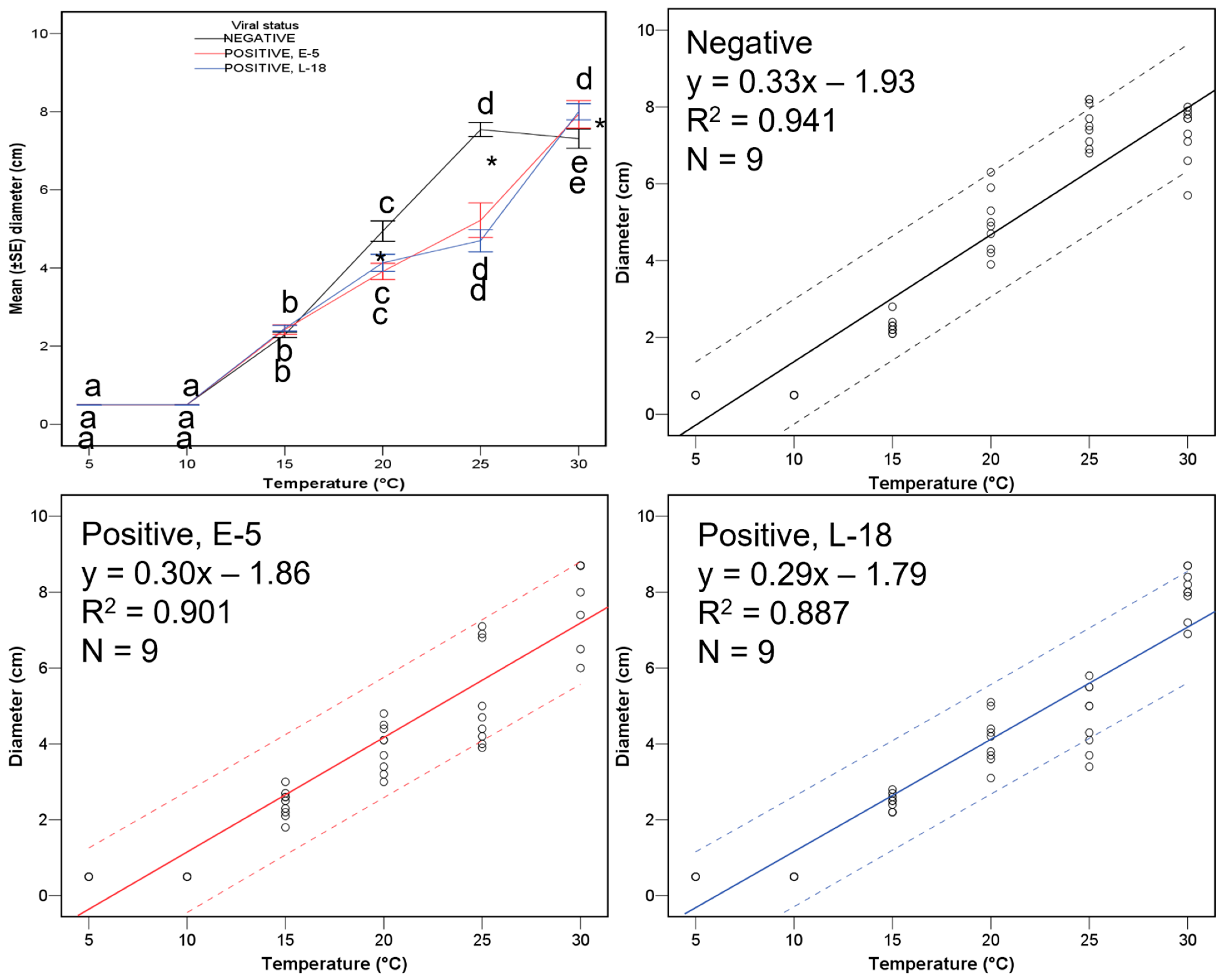
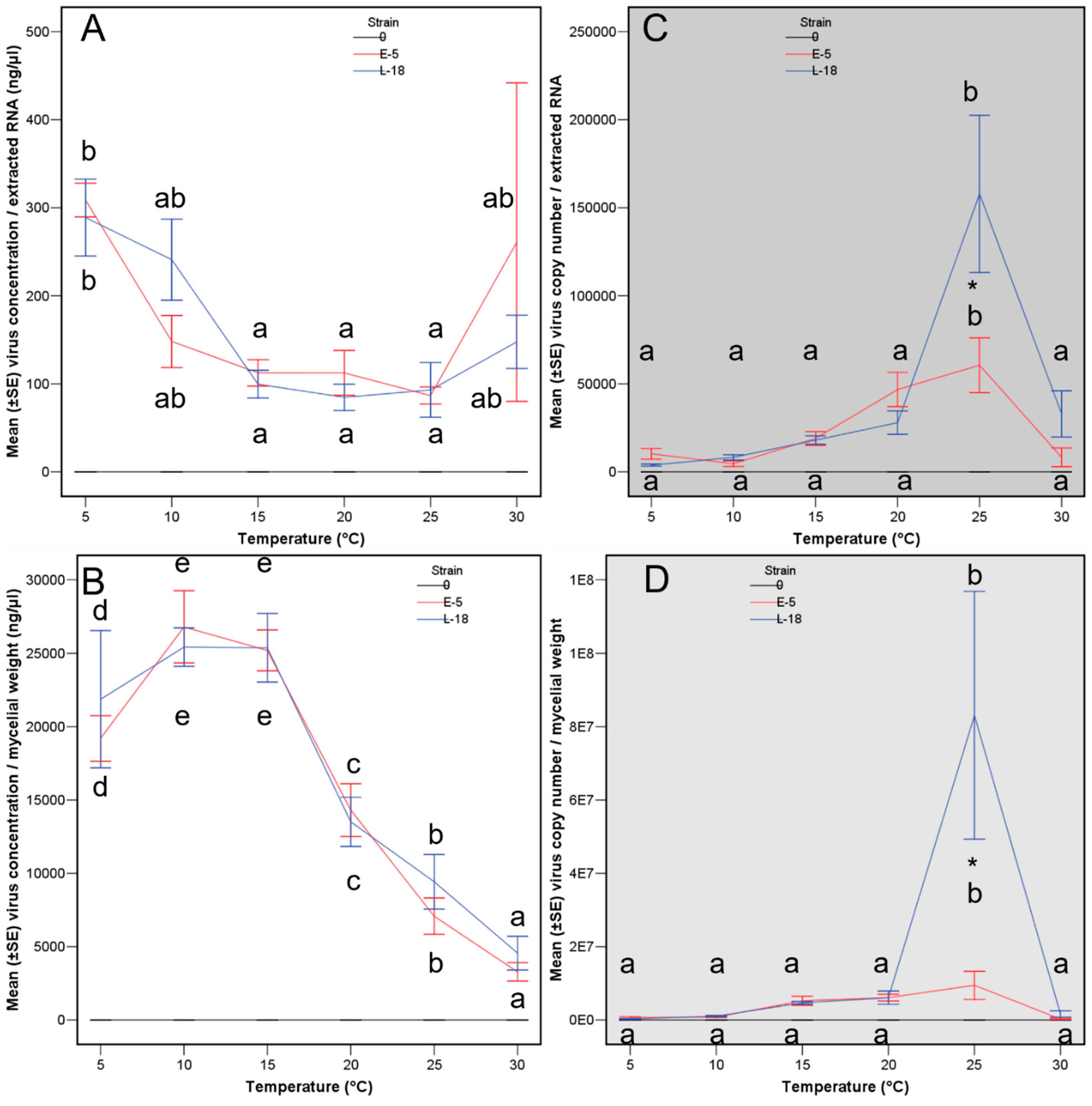
| Treatment Number | Strain | Transmitted from | VCG (vic Genotype) | Mating Type | Virus Haplotype |
|---|---|---|---|---|---|
| 1 | FTC687 | (SDA540 M2273) | EU10 (2122-11) | MAT-2 | E-5 |
| 2 | WAR706 | (WAP125 M2273) | EU9 (2111-11) | MAT-2 | E-5 |
| 3 | POWP709 | (WAP125 M2273) | EU9 (2111-11) | MAT-2 | E-5 |
| 4 | FTC687 | (SDA540 M2357) | EU10 (2122-11) | MAT-2 | L-18 |
| 5 | WAR706 | (WAP125 M2357) | EU9 (2111-11) | MAT-2 | L-18 |
| 6 | POWP709 | (WAP125 M2357) | EU9 (2111-11) | MAT-2 | L-18 |
| 7 | FTC687 NON-INFECTED | Virus-free | EU10 (2122-11) | MAT-2 | N/A |
| 8 | WAR706 NON-INFECTED | Virus-free | EU9 (2111-11) | MAT-2 | N/A |
| 9 | POWP709 NON-INFECTED | Virus-free | EU9 (2111-11) | MAT-2 | N/A |
| Virus Concentration (ng/µL) | Virus Copy Number | |||||||
| Model | Temperature | Fungal Replicate | Viral Strain | Model | Temperature | Fungal Replicate | Viral Strain | |
| Type III Sum of Squares | 6,012,136.81 | 5,978,713.19 | 13,605.38 | 19,818.23 | 685,529,198,694,467.0 | 604,331,700,009,547.0 | 2,224,111,442,904.95 | 78,973,387,242,015 |
| df | 8 | 5 | 2 | 1 | 8 | 5 | 2 | 1 |
| Mean Square | 751,517.10 | 1,195,742.63 | 6802.69 | 19,818.23 | 85,691,149,836,808.40 | 120,866,340,001,909.5 | 1,112,055,721,452.47 | 78,973,387,242,015 |
| Observed Power | 1.00 | 1.00 | 0.06 | 0.08 | 0.99 | 0.99 | 0.06 | 0.58 |
| R2 | 0.50 | 0.50 | 0.50 | 0.50 | 0.29 | 0.29 | 0.29 | 0.29 |
| F | 12.66 | 20.15 | 0.11 | 0.33 | 5.19 | 7.32 | 0.06 | 4.78 |
| p | 0.0001 | 0.0001 | 0.89 | 0.56 | 0.0001 | 0.0001 | 0.93 | 0.03 |
| Virus Concentration/Extracted RNA (ng/µL) | Virus Copy Number/Extracted RNA | |||||||
| Model | Temperature | Fungal Replicate | Viral Strain | Model | Temperature | Fungal Replicate | Viral Strain | |
| Type III Sum of Squares | 699,833.66 | 609,462.93 | 86,195.18 | 4175.54 | 145,068,661,314.46 | 136,296,027,364.27 | 1,366,249,325.82 | 7,406,384,624.36 |
| df | 8 | 5 | 2 | 1 | 8 | 5 | 2 | 1 |
| Mean Square | 87,479.20 | 121,892.58 | 43,097.59 | 4175.54 | 18,133,582,664.30 | 27,259,205,472.85 | 683,124,662.91 | 7,406,384,624.36 |
| Observed Power | 0.93 | 0.94 | 0.30 | 0.06 | 1.00 | 1.00 | 0.09 | 0.43 |
| R2 | 0.19 | 0.19 | 0.19 | 0.19 | 0.39 | 0.39 | 0.39 | 0.39 |
| F | 2.90 | 4.04 | 1.42 | 0.13 | 7.94 | 11.94 | 0.29 | 3.24 |
| p | 0.006 | 0.002 | 0.24 | 0.71 | 0.0001 | 0.0001 | 0.74 | 0.05 |
| Virus Concentration/Mycelial Weight (ng/µL) | Virus Copy Number/Mycelial Weight | |||||||
| Model | Temperature | Fungal Replicate | Viral Strain | Model | Temperature | Fungal Replicate | Viral Strain | |
| Type III Sum of Squares | 7,634,052,101.53 | 7,540,863,471.70 | 79,309,812.22 | 13,878,817.59 | 33,464,956,110,528,400 | 29,013,085,112,384,300 | 321,942,634,010,970.6 | 4,129,928,364,133,164 |
| df | 8 | 5 | 2 | 1 | 8 | 5 | 2 | 1 |
| Mean Square | 954,256,512.69 | 1,508,172,694.34 | 39,654,906.11 | 13,878,817.59 | 4,183,119,513,816,050.00 | 5,802,617,022,476,860.00 | 160,971,317,005,485.30 | 412,992,836,413,3164.00 |
| Observed Power | 1.00 | 1.00 | 0.22 | 0.09 | 0.98 | 0.98 | 0.07 | 0.50 |
| R2 | 0.67 | 0.67 | 0.67 | 0.67 | 0.24 | 0.24 | 0.24 | 0.24 |
| F | 25.25 | 39.90 | 1.04 | 0.36 | 4.00 | 5.56 | 0.15 | 3.95 |
| p | 0.0001 | 0.0001 | 0.35 | 0.54 | 0.0001 | 0.0001 | 0.85 | 0.04 |
| Colony Diameter (Overall) | Colony Diameter (without Negatives) | |||||||
|---|---|---|---|---|---|---|---|---|
| Model | Temperature | Fungal Replicate | Viral Strain | Model | Temperature | Fungal Replicate | Viral Strain | |
| Type III Sum of Squares | 177.93 | 1170.40 | 0.20 | 7.32 | 745.70 | 745.60 | 0.075 | 0.03 |
| df | 9 | 5 | 2 | 2 | 8 | 5 | 2 | 1 |
| Mean Square | 130.88 | 234.08 | 0.10 | 3.66 | 93.21 | 149.12 | 0.037 | 0.03 |
| Observed Power | 1.00 | 1.00 | 0.07 | 0.87 | 1.00 | 1.00 | 0.06 | 0.05 |
| R2 | 0.92 | 0.92 | 0.92 | 0.92 | 0.94 | 0.94 | 0.94 | 0.94 |
| F | 211.43 | 378.14 | 0.16 | 5.91 | 222.94 | 356.66 | 0.08 | 0.07 |
| p | 0.0001 | 0.0001 | 0.84 | 0.003 | 0.0001 | 0.0001 | 0.91 | 0.78 |
| Virus Concentration (ng/µL) | Virus Copy Number | |||||||
|---|---|---|---|---|---|---|---|---|
| Model | Temperature | Fungal Replicate | Viral Strain | Model | Temperature | Fungal Replicate | Viral Strain | |
| Type III Sum of Squares | 1,556,364.86 | 207,684.71 | 81643.77 | 1,267,036.37 | 14,143,441,842.83 | 3,703,425,992.14 | 1,119,836,716.47 | 9,320,179,134.21 |
| df | 8 | 5 | 2 | 1 | 8 | 5 | 2 | 1 |
| Mean Square | 194,545.60 | 41,536.94 | 40,821.88 | 1,267,036.37 | 1,767,930,230.35 | 740,685,198.42 | 559,918,358.24 | 9,320,179,134.21 |
| Observed Power | 1.00 | 0.83 | 0.55 | 1.00 | 1.00 | 0.76 | 0.38 | 1.00 |
| R2 | 0.52 | 0.52 | 0.52 | 0.52 | 0.32 | 0.32 | 0.32 | 0.32 |
| F | 13.74 | 2.93 | 2.88 | 89.54 | 5.94 | 2.49 | 1.88 | 31.33 |
| p | 0.0001 | 0.01 | 0.06 | 0.0001 | 0.0001 | 0.03 | 0.15 | 0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romon-Ochoa, P.; Smith, O.; Lewis, A.; Kupper, Q.; Shamsi, W.; Rigling, D.; Pérez-Sierra, A.; Ward, L. Temperature Effects on the Cryphonectria hypovirus 1 Accumulation and Recovery within Its Fungal Host, the Chestnut Blight Pathogen Cryphonectria parasitica. Viruses 2023, 15, 1260. https://doi.org/10.3390/v15061260
Romon-Ochoa P, Smith O, Lewis A, Kupper Q, Shamsi W, Rigling D, Pérez-Sierra A, Ward L. Temperature Effects on the Cryphonectria hypovirus 1 Accumulation and Recovery within Its Fungal Host, the Chestnut Blight Pathogen Cryphonectria parasitica. Viruses. 2023; 15(6):1260. https://doi.org/10.3390/v15061260
Chicago/Turabian StyleRomon-Ochoa, Pedro, Olivia Smith, Alex Lewis, Quirin Kupper, Wajeeha Shamsi, Daniel Rigling, Ana Pérez-Sierra, and Lisa Ward. 2023. "Temperature Effects on the Cryphonectria hypovirus 1 Accumulation and Recovery within Its Fungal Host, the Chestnut Blight Pathogen Cryphonectria parasitica" Viruses 15, no. 6: 1260. https://doi.org/10.3390/v15061260





