Mpox Virus: Its Molecular Evolution and Potential Impact on Viral Epidemiology
Abstract
:1. Introduction
2. Mpox Virus
2.1. Introduction to the Family Poxviridae
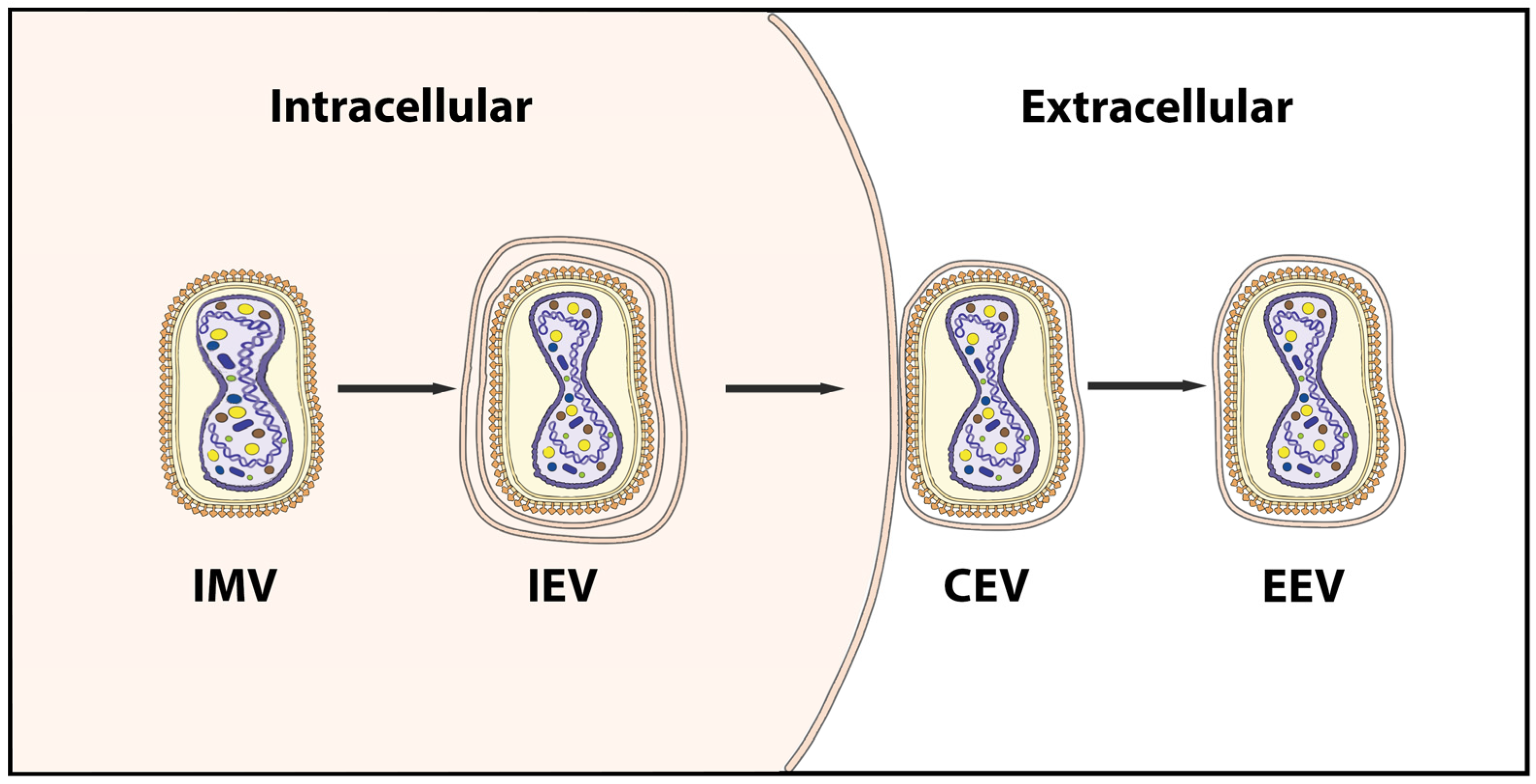
2.2. Genome and Morphology of MPXV
2.3. Hosts and Reservoirs
3. Virus–Host Interaction and Immunology
3.1. Innate Immune Responses to MPXV
3.2. Adaptive Immune Responses to MPXV
3.3. Immune Evasion
4. Transmission and Epidemiology
4.1. Transmission
4.2. Epidemiology
5. Phylogeny and Evolution
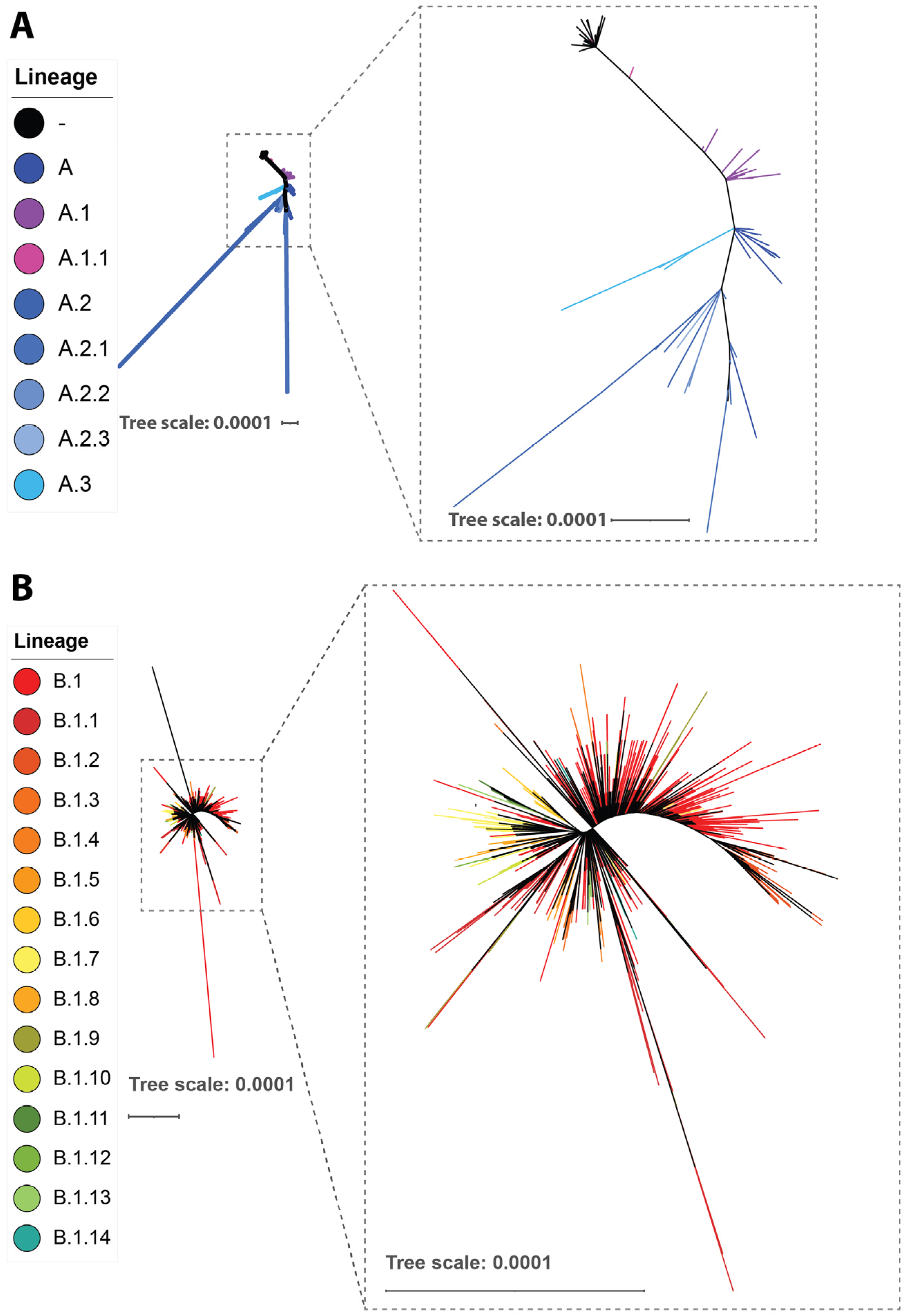
5.1. Phylogeny
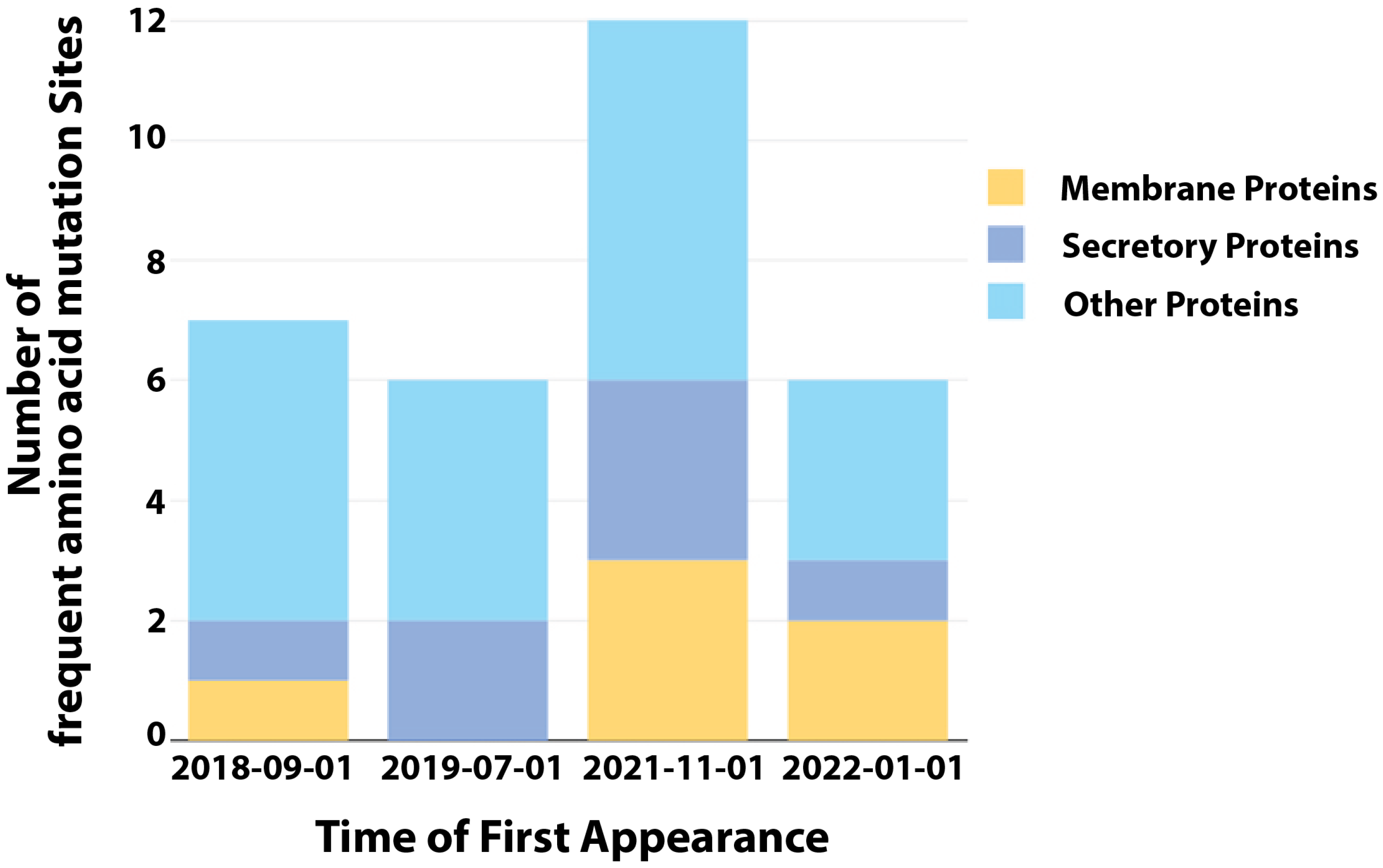
5.2. SNPs
5.3. Recombination
5.4. Gene Loss and Amplification
6. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cho, C.T.; Wenner, H.A. Monkeypox virus. Bacteriol. Rev. 1973, 37, 1–18. [Google Scholar] [CrossRef]
- Mccollum, A.M.; Damon, I.K. Human Monkeypox. Clin. Infect. Dis. 2013, 58, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Jezek, Z.; Szczeniowski, M.; Paluku, K.M.; Mutombo, M. Human Monkeypox: Clinical Features of 282 Patients. J. Infect. Dis. 1987, 156, 293–298. [Google Scholar] [CrossRef] [PubMed]
- CDC Mpox in the U.S. Available online: https://www.cdc.gov/poxvirus/monkeypox/response/2022/world-map.html (accessed on 15 January 2023).
- Otu, A.; Ebenso, B.; Walley, J.; Barceló, J.M.; Ochu, C.L. Global human monkeypox outbreak: Atypical presentation demanding urgent public health action. Lancet Microbe 2022, 3, e554–e555. [Google Scholar] [CrossRef]
- Minhaj, F.S.; Ogale, Y.P.; Whitehill, F.; Schultz, J.; Foote, M.; Davidson, W.; Hughes, C.M.; Wilkins, K.; Bachmann, L.; Chatelain, R.; et al. Monkeypox Outbreak—Nine States, May 2022. MMWR. Morb. Mortal. Wkly. Rep. 2022, 71, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Desingu, P.A.; Nagarajan, K. Genomic Regions Insertion and Deletion in Monkeypox Virus Causing Multi-Country Out-break-2022. bioRxiv 2022. [Google Scholar] [CrossRef]
- Barrett, J.; McFadden, G. Origin and Evolution of Poxviruses, 2nd ed.; Domingo, E., Parrish, C., Holland, J., Eds.; Origin and Evolution of Viruses; Academic Press: Salt Lake City, UT, USA, 2008; p. 446. ISBN 978-0-08-056496-8. [Google Scholar]
- Hughes, A.L.; Irausquin, S.; Friedman, R. The evolutionary biology of poxviruses. Infect. Genet. Evol. 2010, 10, 50–59. [Google Scholar] [CrossRef]
- Schmidt, F.I.; Bleck, C.K.E.; Mercer, J. Poxvirus host cell entry. Curr. Opin. Virol. 2012, 2, 20–27. [Google Scholar] [CrossRef]
- Kilcher, S.; Schmidt, F.I.; Schneider, C.; Kopf, M.; Helenius, A.; Mercer, J. siRNA Screen of Early Poxvirus Genes Identifies the AAA+ ATPase D5 as the Virus Genome-Uncoating Factor. Cell Host Microbe 2014, 15, 103–112. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus entry and membrane fusion. Virology 2006, 344, 48–54. [Google Scholar] [CrossRef]
- Moss, B. Vaccinia Virus: A Tool for Research and Vaccine Development. Science 1991, 252, 1662–1667. [Google Scholar] [CrossRef] [PubMed]
- Payne, L.G. Significance of Extracellular Enveloped Virus in the in vitro and in vivo Dissemination of Vaccinia. J. Gen. Virol. 1980, 50, 89–100. [Google Scholar] [CrossRef]
- Shchelkunov, S.N.; Totmenin, A.V.; Babkin, I.V.; Safronov, P.F.; Ryazankina, O.I.; Petrov, N.A.; Gutorov, V.V.; Uvarova, E.; Mikheev, M.V.; Sisler, J.R.; et al. Human monkeypox and smallpox viruses: Genomic comparison. FEBS Lett. 2001, 509, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Kugelman, J.R.; Johnston, S.C.; Mulembakani, P.M.; Kisalu, N.; Lee, M.S.; Koroleva, G.; McCarthy, S.E.; Gestole, M.C.; Wolfe, N.D.; Fair, J.N.; et al. Genomic Variability of Monkeypox Virus among Humans, Democratic Republic of the Congo. Emerg. Infect. Dis. 2014, 20, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Seet, B.T.; Johnston, J.B.; Brunetti, C.R.; Barrett, J.W.; Everett, H.; Cameron, C.; Sypula, J.; Nazarian, S.H.; Lucas, A.; McFadden, G. Poxviruses and Immune Evasion. Ann. Rev. Immunol. 2003, 21, 377–423. [Google Scholar] [CrossRef]
- Reed, K.D.; Melski, J.W.; Graham, M.B.; Regnery, R.L.; Sotir, M.J.; Wegner, M.V.; Kazmierczak, J.J.; Stratman, E.J.; Li, Y.; Fairley, J.A.; et al. The Detection of Monkeypox in Humans in the Western Hemisphere. N. Engl. J. Med. 2004, 350, 342–350. [Google Scholar] [CrossRef]
- Resch, W.; Hixson, K.K.; Moore, R.J.; Lipton, M.S.; Moss, B. Protein composition of the vaccinia virus mature virion. Virology 2006, 358, 233–247. [Google Scholar] [CrossRef]
- Manes, N.P.; Estep, R.D.; Mottaz, H.M.; Moore, R.J.; Clauss, T.R.W.; Monroe, M.E.; Du, X.; Adkins, J.N.; Wong, S.W.; Smith, R.D. Comparative Proteomics of Human Monkeypox and Vaccinia Intracellular Mature and Extracellular Enveloped Virions. J. Proteome Res. 2008, 7, 960–968. [Google Scholar] [CrossRef]
- Shchelkunov, S.; Totmenin, A.; Safronov, P.; Mikheev, M.; Gutorov, V.; Ryazankina, O.; Petrov, N.; Babkin, I.; Uvarova, E.; Sandakhchiev, L.; et al. Analysis of the Monkeypox Virus Genome. Virology 2002, 297, 172–194. [Google Scholar] [CrossRef]
- Chen, N.; Li, G.; Liszewski, M.K.; Atkinson, J.P.; Jahrling, P.B.; Feng, Z.; Schriewer, J.; Buck, C.; Wang, C.; Lefkowitz, E.J.; et al. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology 2005, 340, 46–63. [Google Scholar] [CrossRef]
- Weaver, J.R.; Isaacs, S.N. Monkeypox virus and insights into its immunomodulatory proteins. Immunol. Rev. 2008, 225, 96–113. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.G.; Damon, I.K. Outbreaks of human monkeypox after cessation of smallpox vaccination. Trends Microbiol. 2012, 20, 80–87. [Google Scholar] [CrossRef] [PubMed]
- McFadden, G. Poxvirus tropism. Nat. Rev. Genet. 2005, 3, 201–213. [Google Scholar] [CrossRef]
- Smith, G.L.; Law, M. The exit of Vaccinia virus from infected cells. Virus Res. 2004, 106, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Pickup, D.J. Extracellular Virions: The Advance Guard of Poxvirus Infections. PLoS Pathog. 2015, 11, e1004904. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, J.-C.; Chung, C.-S.; Chang, W. Vaccinia Virus Envelope D8L Protein Binds to Cell Surface Chondroitin Sulfate and Mediates the Adsorption of Intracellular Mature Virions to Cells. J. Virol. 1999, 73, 8750–8761. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.-S.; Hsiao, J.-C.; Chang, Y.-S.; Chang, W. A27L Protein Mediates Vaccinia Virus Interaction with Cell Surface Heparan Sulfate. J. Virol. 1998, 72, 1577–1585. [Google Scholar] [CrossRef]
- Lin, C.-L.; Chung, C.-S.; Heine, H.; Chang, W. Vaccinia Virus Envelope H3L Protein Binds to Cell Surface Heparan Sulfate and Is Important for Intracellular Mature Virion Morphogenesis and Virus Infection In Vitro and In Vivo. J. Virol. 2000, 74, 3353–3365. [Google Scholar] [CrossRef]
- Moss, B. Membrane fusion during poxvirus entry. Semin. Cell Dev. Biol. 2016, 60, 89–96. [Google Scholar] [CrossRef]
- Khodakevich, L.; Jezek, Z.; Messinger, D. Monkeypox virus—Ecology and public-health significance. Bull. World Health Organ. 1988, 66, 747–752. [Google Scholar]
- Reynolds, M.G.; Suu-Ire, R.; Karem, K.; Root, J.J.; Galley, J.; Carroll, D.S.; Abel, J.; Kwasi, M.O.; Damon, I.K.; Likos, A.; et al. A Silent Enzootic of an Orthopoxvirus in Ghana, West Africa: Evidence for Multi-Species Involvement in the Absence of Widespread Human Disease. Am. J. Trop. Med. Hyg. 2010, 82, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Salzer, J.S.; Carroll, D.S.; Rwego, I.B.; Li, Y.; Falendysz, E.A.; Shisler, J.L.; Karem, K.L.; Damon, I.K.; Gillespie, T.R. Serologic Evidence For Circulating Orthopoxviruses In Peridomestic Rodents From Rural Uganda. J. Wildl. Dis. 2013, 49, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Orba, Y.; Sasaki, M.; Yamaguchi, H.; Ishii, A.; Thomas, Y.; Ogawa, H.; Hang’ombe, B.M.; Mweene, A.S.; Morikawa, S.; Saijo, M.; et al. Orthopoxvirus infection among wildlife in Zambia. J. Gen. Virol. 2015, 96, 390–394. [Google Scholar] [CrossRef]
- Doty, J.B.; Malekani, J.M.; Kalemba, L.N.; Stanley, W.T.; Monroe, B.P.; Nakazawa, Y.U.; Mauldin, M.R.; Bakambana, T.L.; Liyandja, T.L.D.; Braden, Z.H.; et al. Assessing Monkeypox Virus Prevalence in Small Mammals at the Human–Animal Interface in the Democratic Republic of the Congo. Viruses 2017, 9, 283. [Google Scholar] [CrossRef] [PubMed]
- Hutin, Y.J.; Williams, R.J.; Malfait, P.; Pebody, R.; Loparev, V.N.; Ropp, S.L.; Rodriguez, M.; Knight, J.C.; Tshioko, F.K.; Khan, A.S.; et al. Outbreak of Human Monkeypox, Democratic Republic of Congo, 1996 to 1997. Emerg. Infect. Dis. 2001, 7, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Hutson, C.L.; Olson, V.A.; Carroll, D.S.; Abel, J.A.; Hughes, C.M.; Braden, Z.H.; Weiss, S.; Self, J.; Osorio, J.E.; Hudson, P.N.; et al. A prairie dog animal model of systemic orthopoxvirus disease using West African and Congo Basin strains of monkeypox virus. J. Gen. Virol. 2009, 90, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Yinka-Ogunleye, A.; Aruna, O.; Dalhat, M.; Ogoina, D.; McCollum, A.; Disu, Y.; Mamadu, I.; Akinpelu, A.; Ahmad, A.; Burga, J.; et al. Outbreak of human monkeypox in Nigeria in 2017–18: A clinical and epidemiological report. Lancet Infect. Dis. 2019, 19, 872–879. [Google Scholar] [CrossRef]
- Falendysz, E.A.; Lopera, J.G.; Doty, J.B.; Nakazawa, Y.; Crill, C.; Lorenzsonn, F.; Kalemba, L.N.; Ronderos, M.D.; Mejia, A.; Malekani, J.M.; et al. Characterization of Monkeypox virus infection in African rope squirrels (Funisciurus sp.). PLoS Neglected Trop. Dis. 2017, 11, e0005809. [Google Scholar] [CrossRef]
- Keasey, S.; Pugh, C.; Tikhonov, A.; Chen, G.; Schweitzer, B.; Nalca, A.; Ulrich, R.G. Proteomic Basis of the Antibody Response to Monkeypox Virus Infection Examined in Cynomolgus Macaques and a Comparison to Human Smallpox Vaccination. PLoS ONE 2010, 5, e15547. [Google Scholar] [CrossRef]
- Reynolds, M.G.; Doty, J.B.; Mccollum, A.M.; Olson, V.A.; Nakazawa, Y. Monkeypox re-emergence in Africa: A call to expand the concept and practice of One Health. Expert Rev. Anti-infective Ther. 2018, 17, 129–139. [Google Scholar] [CrossRef]
- Zaucha, G.M.; Jahrling, P.B.; Geisbert, T.W.; Swearengen, J.R.; Hensley, L. The Pathology of Experimental Aerosolized Monkeypox Virus Infection in Cynomolgus Monkeys (Macaca fascicularis). Lab. Investig. 2001, 81, 1581–1600. [Google Scholar] [CrossRef] [PubMed]
- Hammarlund, E.; Dasgupta, A.; Pinilla, C.; Norori, P.; Früh, K.; Slifka, M.K. Monkeypox virus evades antiviral CD4 + and CD8 + T cell responses by suppressing cognate T cell activation. Proc. Natl. Acad. Sci. USA 2008, 105, 14567–14572. [Google Scholar] [CrossRef] [PubMed]
- Rubins, K.H.; Hensley, L.E.; Jahrling, P.B.; Whitney, A.R.; Geisbert, T.W.; Huggins, J.W.; Owen, A.; LeDuc, J.W.; Brown, P.O.; Relman, D.A. The host response to smallpox: Analysis of the gene expression program in peripheral blood cells in a nonhuman primate model. Proc. Natl. Acad. Sci. USA 2004, 101, 15190–15195. [Google Scholar] [CrossRef] [PubMed]
- Jahrling, P.B.; Hensley, L.E.; Martinez, M.J.; LeDuc, J.W.; Rubins, K.H.; Relman, D.A.; Huggins, J.W. Exploring the potential of variola virus infection of cynomolgus macaques as a model for human smallpox. Proc. Natl. Acad. Sci. USA 2004, 101, 15196–15200. [Google Scholar] [CrossRef]
- Rubins, K.H.; Hensley, L.E.; Relman, D.A.; Brown, P.O. Stunned Silence: Gene Expression Programs in Human Cells Infected with Monkeypox or Vaccinia Virus. PLoS ONE 2011, 6, e15615. [Google Scholar] [CrossRef] [PubMed]
- Paust, S.; Senman, B.; Von Andrian, U.H. Adaptive immune responses mediated by natural killer cells. Immunol. Rev. 2010, 235, 286–296. [Google Scholar] [CrossRef]
- Song, H.; Josleyn, N.; Janosko, K.; Skinner, J.; Reeves, R.K.; Cohen, M.; Jett, C.; Johnson, R.; Blaney, J.E.; Bollinger, L.; et al. Monkeypox Virus Infection of Rhesus Macaques Induces Massive Expansion of Natural Killer Cells but Suppresses Natural Killer Cell Functions. PLoS ONE 2013, 8, e77804. [Google Scholar] [CrossRef]
- Johnston, S.C.; Lin, K.L.; Connor, J.H.; Ruthel, G.; Goff, A.; Hensley, L.E. In vitro inhibition of monkeypox virus production and spread by Interferon-β. Virol. J. 2012, 9, 5. [Google Scholar] [CrossRef]
- Johnston, S.C.; Johnson, J.C.; Stonier, S.W.; Lin, K.L.; Kisalu, N.K.; Hensley, L.E.; Rimoin, A.W. Cytokine modulation correlates with severity of monkeypox disease in humans. J. Clin. Virol. 2015, 63, 42–45. [Google Scholar] [CrossRef]
- Liu, L.; Xu, Z.; Fuhlbrigge, R.C.; Peña-Cruz, V.; Lieberman, J.; Kupper, T.S. Vaccinia Virus Induces Strong Immunoregulatory Cytokine Production in Healthy Human Epidermal Keratinocytes: A Novel Strategy for Immune Evasion. J. Virol. 2005, 79, 7363–7370. [Google Scholar] [CrossRef]
- Howell, M.D.; Gallo, R.L.; Boguniewicz, M.; Jones, J.F.; Wong, C.; Streib, J.E.; Leung, D.Y. Cytokine Milieu of Atopic Dermatitis Skin Subverts the Innate Immune Response to Vaccinia Virus. Immunity 2006, 24, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Broek, M.V.D.; Bachmann, M.F.; Köhler, G.; Barner, M.; Escher, R.; Zinkernagel, R.; Kopf, M. IL-4 and IL-10 Antagonize IL-12-Mediated Protection Against Acute Vaccinia Virus Infection with a Limited Role of IFN-γ and Nitric Oxide Synthetase 2. J. Immunol. 2000, 164, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Mikkelsen, H.; Jungersen, G. Intracellular Pathogens: Host Immunity and Microbial Persistence Strategies. J. Immunol. Res. 2019, 2019, 1356540. [Google Scholar] [CrossRef] [PubMed]
- Strassburg, M.A. The global eradication of smallpox. Am. J. Infect. Control. 1982, 10, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.D.; McIntosh, K.; Connor, J.D.; Benenson, A.S.; Alling, D.W.; Rolfe, U.T.; Todd, W.A.; Schanberger, J.E.; Mattheis, M.J. Primary Percutaneous Vaccination. J. Infect. Dis. 1977, 135, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Edghill-Smith, Y.; Golding, H.; Manischewitz, J.; King, L.R.; Scott, D.; Bray, M.; Nalca, A.; Hooper, J.; Whitehouse, C.A.; Schmitz, J.E.; et al. Smallpox vaccine–induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat. Med. 2005, 11, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, B.L.; Langland, J.O.; Kibler, K.V.; Denzler, K.L.; White, S.D.; Holechek, S.A.; Wong, S.; Huynh, T.; Baskin, C.R. Vaccinia virus vaccines: Past, present and future. Antivir. Res. 2009, 84, 1–13. [Google Scholar] [CrossRef]
- Crotty, S.; Felgner, P.; Davies, H.; Glidewell, J.; Villarreal, L.; Ahmed, R. Cutting Edge: Long-Term B Cell Memory in Humans after Smallpox Vaccination. J. Immunol. 2003, 171, 4969–4973. [Google Scholar] [CrossRef]
- Hammarlund, E.; Lewis, M.W.; Hansen, S.G.; Strelow, L.I.; Nelson, J.A.; Sexton, G.J.; Hanifin, J.M.; Slifka, M.K. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 2003, 9, 1131–1137. [Google Scholar] [CrossRef]
- Mack, T.M.; Noble, J., Jr.; Thomas, D.B. A Prospective Study of Serum Antibody and Protection Against Smallpox. Am. J. Trop. Med. Hyg. 1972, 21, 214–218. [Google Scholar] [CrossRef]
- MacLeod, M.K.; Clambey, E.T.; Kappler, J.W.; Marrack, P. CD4 memory T cells: What are they and what can they do? Semin. Immunol. 2009, 21, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Hickman, H.D.; Reynoso, G.V.; Ngudiankama, B.F.; Rubin, E.J.; Magadán, J.G.; Cush, S.S.; Gibbs, J.; Molon, B.; Bronte, V.; Bennink, J.R.; et al. Anatomically Restricted Synergistic Antiviral Activities of Innate and Adaptive Immune Cells in the Skin. Cell Host Microbe 2013, 13, 155–168. [Google Scholar] [CrossRef]
- Marco, M.D.M.F.D.; Alejo, A.; Hudson, P.; Damon, I.K.; Alcami, A. The highly virulent variola and monkeypox viruses express secreted inhibitors of type I interferon. FASEB J. 2009, 24, 1479–1488. [Google Scholar] [CrossRef]
- Esteban, D.J.; Nuara, A.A.; Buller, R.M.L. Interleukin-18 and glycosaminoglycan binding by a protein encoded by Variola virus. J. Gen. Virol. 2004, 85, 1291–1299. [Google Scholar] [CrossRef]
- Takata, M.A.; Gonçalves-Carneiro, D.; Zang, T.M.; Soll, S.J.; York, A.; Blanco-Melo, D.; Bieniasz, P.D. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 2017, 550, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, D.; Nchioua, R.; Sherrill-Mix, S.; Stürzel, C.M.; Heusinger, E.; Braun, E.; Gondim, M.V.P.; Hotter, D.; Sparrer, K.M.J.; Hahn, B.H.; et al. CpG Frequency in the 5′ Third of the env Gene Determines Sensitivity of Primary HIV-1 Strains to the Zinc-Finger Antiviral Protein. Mbio 2020, 11, e02903-19. [Google Scholar] [CrossRef] [PubMed]
- Nchioua, R.; Kmiec, D.; Müller, J.A.; Conzelmann, C.; Groß, R.; Swanson, C.M.; Neil, S.J.D.; Stenger, S.; Sauter, D.; Münch, J.; et al. SARS-CoV-2 Is Restricted by Zinc Finger Antiviral Protein despite Preadaptation to the Low-CpG Environment in Humans. mBio 2020, 11, e01930-20. [Google Scholar] [CrossRef]
- Peng, C.; Wyatt, L.S.; Glushakow-Smith, S.G.; Lal-Nag, M.; Weisberg, A.S.; Moss, B. Zinc-finger antiviral protein (ZAP) is a restriction factor for replication of modified vaccinia virus Ankara (MVA) in human cells. PLoS Pathog. 2020, 16, e1008845. [Google Scholar] [CrossRef]
- Hudson, P.N.; Self, J.; Weiss, S.; Braden, Z.; Xiao, Y.; Girgis, N.M.; Emerson, G.; Hughes, C.; Sammons, S.A.; Isaacs, S.N.; et al. Elucidating the Role of the Complement Control Protein in Monkeypox Pathogenicity. PLoS ONE 2012, 7, e35086. [Google Scholar] [CrossRef]
- Bunge, E.M.; Hoet, B.; Chen, L.; Lienert, F.; Weidenthaler, H.; Baer, L.R.; Steffen, R. The changing epidemiology of human monkeypox—A potential threat? A systematic review. PLoS Neglected Trop. Dis. 2022, 16, e0010141. [Google Scholar] [CrossRef]
- Nolen, L.D.; Tamfum, J.-J.M.; Kabamba, J.; Likofata, J.; Katomba, J.; McCollum, A.M.; Monroe, B.; Kalemba, L.; Mukadi, D.; Bomponda, P.L.; et al. Introduction of Monkeypox into a Community and Household: Risk Factors and Zoonotic Reservoirs in the Democratic Republic of the Congo. Am. J. Trop. Med. Hyg. 2015, 93, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.G.; Davidson, W.B.; Curns, A.T.; Conover, C.S.; Huhn, G.; Davis, J.P.; Wegner, M.; Croft, D.R.; Newman, A.; Obiesie, N.N.; et al. Spectrum of Infection and Risk Factors for Human Monkeypox, United States, 2003. Emerg. Infect. Dis. 2007, 13, 1332–1339. [Google Scholar] [CrossRef] [PubMed]
- HAN Archive—00446 | Health Alert Network (HAN). Available online: https://emergency.cdc.gov/han/2021/han00446.asp (accessed on 15 January 2023).
- Sklenovská, N.; Van Ranst, M. Emergence of Monkeypox as the Most Important Orthopoxvirus Infection in Humans. Front. Public Heal. 2018, 6, 241. [Google Scholar] [CrossRef] [PubMed]
- Von Magnus, P.; Andersen, E.K.; Petersen, K.B.; Birch-Andersen, A. A pox-like disease in cynomolgus monkeys. Acta Pathol. Microbiol. Scand. 2009, 46, 156–176. [Google Scholar] [CrossRef]
- Marennikova, S.S.; Seluhina, E.M.; Mal’Ceva, N.N.; Cimiskjan, K.L.; Macevic, G.R. Isolation and properties of the causal agent of a new variola-like disease (monkeypox) in man. Bull. World Health Organ. 1972, 46, 599–611. [Google Scholar]
- Ladnyj, I.; Ziegler, P.; Kima, E. Human infection caused by monkeypox virus in basankusu terri-tory, democratic-republic-of-congo. Bull. World Health Organ. 1972, 46, 593. [Google Scholar]
- Foster, S.; Eke, R.; Foege, W.; Smith, E.; Titus, J.; Moser, C.; Lourie, B.; Brink, E.; Kuteyi, O.; Cummings, E.; et al. HUMAN MONKEYPOX. Bull. World Health Organ. 1972, 46, 569–576. [Google Scholar]
- Huhn, G.D.; Bauer, A.M.; Yorita, K.; Graham, M.B.; Sejvar, J.; Likos, A.; Damon, I.K.; Reynolds, M.; Kuehnert, M.J. Clinical Characteristics of Human Monkeypox, and Risk Factors for Severe Disease. Clin. Infect. Dis. 2005, 41, 1742–1751. [Google Scholar] [CrossRef]
- Fleischauer, A.T.; Kile, J.C.; Davidson, M.; Fischer, M.; Karem, K.L.; Teclaw, R.; Messersmith, H.; Pontones, P.; Beard, B.A.; Braden, Z.H.; et al. Evaluation of Human-to-Human Transmission of Monkeypox from Infected Patients to Health Care Workers. Clin. Infect. Dis. 2005, 40, 689–694. [Google Scholar] [CrossRef]
- Adler, H.; Gould, S.; Hine, P.; Snell, L.B.; Wong, W.; Houlihan, C.F.; Osborne, J.C.; Rampling, T.; Beadsworth, M.B.; Duncan, C.J.; et al. Clinical features and management of human monkeypox: A retrospective observational study in the UK. Lancet Infect. Dis. 2022, 22, 1153–1162. [Google Scholar] [CrossRef]
- Bragazzi, N.L.; Kong, J.D.; Mahroum, N.; Tsigalou, C.; Khamisy-Farah, R.; Converti, M.; Wu, J. Epidemiological trends and clinical features of the ongoing monkeypox epidemic: A preliminary pooled data analysis and literature review. J. Med. Virol. 2022, 95, e27931. [Google Scholar] [CrossRef] [PubMed]
- Multi-Country Monkeypox Outbreak: Situation Update. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON390 (accessed on 15 January 2023).
- Nguyen, P.-Y.; Ajisegiri, W.S.; Costantino, V.; Chughtai, A.A.; MacIntyre, C.R. Reemergence of Human Monkeypox and Declining Population Immunity in the Context of Urbanization, Nigeria, 2017–2020. Emerg. Infect. Dis. 2021, 27, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.; Kantele, A.; Koopmans, M.; Asogun, D.; Yinka-Ogunleye, A.; Ihekweazu, C.; Zumla, A. Human monkeypox: Epidemiologic and clinical characteristics, diagnosis, and prevention. Infect. Dis. Clin. 2019, 33, 1027–1043. [Google Scholar] [CrossRef] [PubMed]
- Simpson, K.; Heymann, D.; Brown, C.S.; Edmunds, W.J.; Elsgaard, J.; Fine, P.; Hochrein, H.; Hoff, N.A.; Green, A.; Ihekweazu, C.; et al. Human monkeypox—After 40 years, an unintended consequence of smallpox eradication. Vaccine 2020, 38, 5077–5081. [Google Scholar] [CrossRef]
- Reynolds, M.G.; Carroll, D.S.; Karem, K.L. Factors affecting the likelihood of monkeypox’s emergence and spread in the post-smallpox era. Curr. Opin. Virol. 2012, 2, 335–343. [Google Scholar] [CrossRef]
- Likos, A.M.; Sammons, S.A.; Olson, V.A.; Frace, A.M.; Li, Y.; Olsen-Rasmussen, M.; Davidson, W.; Galloway, R.; Khristova, M.L.; Reynolds, M.G.; et al. A tale of two clades: Monkeypox viruses. J. Gen. Virol. 2005, 86, 2661–2672. [Google Scholar] [CrossRef]
- Beer, E.M.; Rao, V.B. A systematic review of the epidemiology of human monkeypox outbreaks and implications for outbreak strategy. PLoS Neglected Trop. Dis. 2019, 13, e0007791. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2006, 23, 127–128. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, A.; Aarons, E.; Astbury, J.; Balasegaram, S.; Beadsworth, M.; Beck, C.R.; Chand, M.; O’connor, C.; Dunning, J.; Ghebrehewet, S.; et al. Two cases of monkeypox imported to the United Kingdom, September 2018. Eurosurveillance 2018, 23, 1800509. [Google Scholar] [CrossRef]
- Erez, N.; Achdout, H.; Milrot, E.; Schwartz, Y.; Wiener-Well, Y.; Paran, N.; Politi, B.; Tamir, H.; Israely, T.; Weiss, S.; et al. Diagnosis of Imported Monkeypox, Israel, 2018. Emerg. Infect. Dis. 2019, 25, 980–983. [Google Scholar] [CrossRef]
- Yong, S.E.F.; Ng, O.T.; Ho, Z.J.M.; Mak, T.M.; Marimuthu, K.; Vasoo, S.; Yeo, T.W.; Ng, Y.K.; Cui, L.; Ferdous, Z.; et al. Imported Monkeypox, Singapore. Emerg. Infect. Dis. 2020, 26, 1826–1830. [Google Scholar] [CrossRef]
- Vaughan, A.; Aarons, E.; Astbury, J.; Brooks, T.; Chand, M.; Flegg, P.; Hardman, A.; Harper, N.; Jarvis, R.; Mawdsley, S.; et al. Human-to-Human Transmission of Monkeypox Virus, United Kingdom, October 2018. Emerg. Infect. Dis. 2020, 26, 782–785. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010199. [Google Scholar] [CrossRef]
- Firth, C.; Kitchen, A.; Shapiro, B.; Suchard, M.A.; Holmes, E.C.; Rambaut, A. Using Time-Structured Data to Estimate Evolutionary Rates of Double-Stranded DNA Viruses. Mol. Biol. Evol. 2010, 27, 2038–2051. [Google Scholar] [CrossRef]
- Duggan, A.T.; Perdomo, M.F.; Piombino-Mascali, D.; Marciniak, S.; Poinar, D.; Emery, M.V.; Buchmann, J.P.; Duchêne, S.; Jankauskas, R.; Humphreys, M.; et al. 17th Century Variola Virus Reveals the Recent History of Smallpox. Curr. Biol. 2016, 26, 3407–3412. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Isidro, J.; Borges, V.; Pinto, M.; Sobral, D.; Santos, J.D.; Nunes, A.; Mixão, V.; Ferreira, R.; Santos, D.; Duarte, S.; et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat. Med. 2022, 28, 1569–1572. [Google Scholar] [CrossRef] [PubMed]
- Jern, P.; Russell, R.A.; Pathak, V.; Coffin, J.M. Likely Role of APOBEC3G-Mediated G-to-A Mutations in HIV-1 Evolution and Drug Resistance. PLoS Pathog. 2009, 5, e1000367. [Google Scholar] [CrossRef] [PubMed]
- Martinez, T.; Shapiro, M.; Bhaduri-McIntosh, S.; MacCarthy, T. Evolutionary effects of the AID/APOBEC family of mutagenic enzymes on human gamma-herpesviruses. Virus Evol. 2019, 5, vey040. [Google Scholar] [CrossRef]
- Kremer, M.; Suezer, Y.; Martinez-Fernandez, Y.; Münk, C.; Sutter, G.; Schnierle, B.S. Vaccinia virus replication is not affected by APOBEC3 family members. Virol. J. 2006, 3, 86. [Google Scholar] [CrossRef]
- Giorgi, F.M.; Pozzobon, D.; Meglio, A.D.; Mercatelli, D. Genomic Analysis of the Recent Monkeypox Outbreak. Med 2022, 3, 824–826. [Google Scholar]
- Gubser, C.; Hué, S.; Kellam, P.; Smith, G.L. Poxvirus genomes: A phylogenetic analysis. J. Gen. Virol. 2004, 85, 105–117. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Arenas, M.; Galán, J.C.; Palero, F.; González-Candelas, F. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol. 2015, 30, 296–307. [Google Scholar] [CrossRef]
- Esposito, J.J.; Sammons, S.A.; Frace, A.M.; Osborne, J.D.; Olsen-Rasmussen, M.; Zhang, M.; Govil, D.; Damon, I.K.; Kline, R.; Laker, M.; et al. Genome Sequence Diversity and Clues to the Evolution of Variola (Smallpox) Virus. Science 2006, 313, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Coulson, D.; Upton, C. Characterization of indels in poxvirus genomes. Virus Genes 2010, 42, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.H. Poxvirus Recombination. Pathogens 2022, 11, 896. [Google Scholar] [CrossRef] [PubMed]
- Sasani, T.A.; Cone, K.R.; Quinlan, A.R.; Elde, N.C. Long read sequencing reveals poxvirus evolution through rapid homogenization of gene arrays. Elife 2018, 7, e35453. [Google Scholar] [CrossRef] [PubMed]
- Cone, K.R.; Kronenberg, Z.N.; Yandell, M.; Elde, N.C. Emergence of a Viral RNA Polymerase Variant during Gene Copy Number Amplification Promotes Rapid Evolution of Vaccinia Virus. J. Virol. 2017, 91, e01428-16. [Google Scholar] [CrossRef] [PubMed]
- Yeh, T.-Y.; Hsieh, Z.-Y.; Feehley, M.C.; Feehley, P.J.; Contreras, G.P.; Su, Y.-C.; Hsieh, S.-L.; Lewis, D.A. Recombination shapes the 2022 monkeypox (mpox) outbreak. Med 2022, 3, 824–826. [Google Scholar] [CrossRef]
- Estep, R.D.; Messaoudi, I.; O’Connor, M.A.; Li, H.; Sprague, J.; Barron, A.; Engelmann, F.; Yen, B.; Powers, M.F.; Jones, J.M.; et al. Deletion of the Monkeypox Virus Inhibitor of Complement Enzymes Locus Impacts the Adaptive Immune Response to Monkeypox Virus in a Nonhuman Primate Model of Infection. J. Virol. 2011, 85, 9527–9542. [Google Scholar] [CrossRef] [PubMed]
- Goff, A.; Mucker, E.; Raymond, J.; Fisher, R.; Bray, M.; Hensley, L.; Paragas, J. Infection of cynomolgus macaques with a recombinant monkeypox virus encoding green fluorescent protein. Arch. Virol. 2011, 156, 1877–1881. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Friedman, R. Poxvirus genome evolution by gene gain and loss. Mol. Phylogenet. Evol. 2005, 35, 186–195. [Google Scholar] [CrossRef]
- Elde, N.C.; Child, S.J.; Eickbush, M.T.; Kitzman, J.O.; Rogers, K.S.; Shendure, J.; Geballe, A.P.; Malik, H.S. Poxviruses Deploy Genomic Accordions to Adapt Rapidly against Host Antiviral Defenses. Cell 2012, 150, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, R.C.; Wang, C.; Hatcher, E.L.; Lefkowitz, E.J. Orthopoxvirus Genome Evolution: The Role of Gene Loss. Viruses 2010, 2, 1933–1967. [Google Scholar] [CrossRef]
- Meyer, H.; Sutter, G.; Mayr, A. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J. Gen. Virol. 1991, 72, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Hatch, G.J.; Graham, V.A.; Bewley, K.R.; Tree, J.A.; Dennis, M.; Taylor, I.; Funnell, S.G.P.; Bate, S.R.; Steeds, K.; Tipton, T.; et al. Assessment of the Protective Effect of Imvamune and Acam2000 Vaccines against Aerosolized Monkeypox Virus in Cynomolgus Macaques. J. Virol. 2013, 87, 7805–7815. [Google Scholar] [CrossRef]
- Parker, S.; Buller, R.M. A review of experimental and natural infections of animals with monkeypox virus between 1958 and 2012. Future Virol. 2013, 8, 129–157. [Google Scholar] [CrossRef]
- Hatcher, E.L.; Hendrickson, R.C.; Lefkowitz, E.J. Identification of Nucleotide-Level Changes Impacting Gene Content and Genome Evolution in Orthopoxviruses. J. Virol. 2014, 88, 13651–13668. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, A.; Kohl, C.; Pape, K.; Bourquain, D.; Thürmer, A.; Michel, J.; Schaade, L.; Nitsche, A. Possible Adaption of the 2022 Monkeypox Virus to the Human Host through Gene Duplication and Loss. bioRxiv 2022. [Google Scholar] [CrossRef]
- Yang, Z. Monkeypox: A potential global threat? J. Med. Virol. 2022, 94, 4034–4036. [Google Scholar] [CrossRef]
- Fine, P.E.M.; Jezek, Z.; Grab, B.; Dixon, H. The Transmission Potential of Monkeypox Virus in Human Populations. Leuk. Res. 1988, 17, 643–650. [Google Scholar] [CrossRef]
- Fenner, F. The Global Eradication Of Smallpox. Med. J. Aust. 1980, 1, 455–456. [Google Scholar] [CrossRef]
- Karim, S.S.A.; Karim, Q.A. Omicron SARS-CoV-2 variant: A new chapter in the COVID-19 pandemic. Lancet. 2021, 398, 2126–2128. [Google Scholar] [CrossRef]
- Sharma, A.; Ahmad Farouk, I.; Lal, S.K. COVID-19: A Review on the Novel Coronavirus Disease Evolution, Transmission, Detection, Control and Prevention. Viruses 2021, 13, 202. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Du, S.; Shan, C.; Nie, K.; Zhang, R.; Li, X.-F.; Zhang, R.; Wang, T.; Qin, C.-F.; et al. Evolutionary enhancement of Zika virus infectivity in Aedes aegypti mosquitoes. Nature 2017, 545, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Shan, C.; Zhu, Y.; Ma, E.; Wang, J.; Wang, P.; Shi, P.-Y.; Cheng, G. A mutation-mediated evolutionary adaptation of Zika virus in mosquito and mammalian host. Proc. Natl. Acad. Sci. USA 2021, 118, e2113015118. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Cheng, G. Contribution of phylogenetics to understanding the evolution and epidemiology of dengue virus. Anim. Model. Exp. Med. 2022, 5, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Cheng, G. Adaptive Evolution as a Driving Force of the Emergence and Re-Emergence of Mosquito-Borne Viral Diseases. Viruses 2022, 14, 435. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, X.; Guo, X.; Peng, W.; Zhu, Y.; Wang, Z.; Yu, X.; Shi, H.; Li, Y.; Zhang, L.; et al. Neighboring mutation-mediated enhancement of dengue virus infectivity and spread. EMBO Rep. 2022, 23, e55671. [Google Scholar] [CrossRef] [PubMed]
| Protein Function | Gene * |
|---|---|
| Ankyrin-like protein | OGP037 |
| CC chemokine binding protein | OPG001 |
| TNF and chemokine binding protein, CrmB | OPG002 |
| Ankyrin-like protein | OPG003 |
| Ankyrin-like protein | OPG003 |
| Ankyrin-like protein | OPG015 |
| Ankyrin-like protein | OPG015 |
| OMCP, inhibitor of natural killer cell-mediated NKG2D-dependent cell lysis | OPG016 |
| Viral growth factor; EGF-like protein | OPG019 |
| Apoptosis inhibitor | OPG021 |
| IL-18 binding protein | OPG022 |
| Ankyrin-like protein | OPG023 |
| Ankyrin-like protein | OPG025 |
| Inhibitor of IRF3 and IRF7 activation; BCL-2-like protein | OPG029 |
| Inhibitor of IRF3 and NF-κB activation, apoptosis inhibitor; BCL-2-like protein | OPG035 |
| Ankyrin-like protein | OPG039 |
| Inhibitor of IRF3 NF-κB activation; BCL-2-like protein | OPG044 |
| Apoptosis inhibitor | OPG045 |
| Double-stranded RNA-binding protein, inhibitor of interferon signalling, apoptosis inhibitor | OPG065 |
| Dephosphorylation of STAT1; phosphatase | OPG106 |
| Inhibitor of MHC class II antigen presentation | OPG163 |
| CC and CXC chemokine binding protein | OPG170 |
| Inhibitor of NF-κB activation; BCL-2-like protein | OPG176 |
| Ankyrin-like protein | OPG189 |
| IFNγ binding proteins | OPG193 |
| Inhibitor of intracellular trafficking of MHC class I molecules | OPG195 |
| Apoptosis inhibitor, caspase 1 and caspase 8 inhibitor, SPI-2 | OPG199 |
| Inhibitor of NF-κB activation; BCL-2-like protein | OPG200 |
| IFNα/β binding proteins | OPG204 |
| Ankyrin-like protein | OPG205 |
| Apoptosis inhibitor, SPI-1 | OPG208 |
| Protein | Sub-Virion Location | Site | Mutation | Time of First Appearance |
|---|---|---|---|---|
| A14L | Membrane | 17 | A17T | 1 September 2018 |
| A19R | Other | 435 | E435K | 1 September 2018 |
| B9R | Secretory | 263 | L263F | 1 September 2018 |
| E6R | Other | 606 | K606E | 1 September 2018 |
| G8L | Other | 196 | D196N | 1 September 2018 |
| H4L | Other | 740 | H740Y | 1 September 2018 |
| L6R | Other | 734 | S734L | 1 September 2018 |
| A19R | Other | 62 | E62K | 1 July 2019 |
| A24R | Other | 307 | S307L | 1 July 2019 |
| J1L | Secretory | 105 | S105L | 1 July 2019 |
| J1R | Other | 264 | D264N | 1 July 2019 |
| J3L | Other | 264 | D264N | 1 July 2019 |
| J3R | Secretory | 105 | S105L | 1 July 2019 |
| A19R | Other | 243 | R243Q | 1 November 2021 |
| B21R | Other | 209 | D209N | 1 November 2021 |
| B21R | Other | 1741 | M1741I | 1 November 2021 |
| C15L | Membrane | 78 | P78S | 1 November 2021 |
| C18L | Membrane | 125 | E125K | 1 November 2021 |
| C9L | Other | 48 | R48C | 1 November 2021 |
| D9L | Secretory | 423 | A423D | 1 November 2021 |
| F8L | Other | 108 | L108F | 1 November 2021 |
| F9R | Membrane | 56 | D56N | 1 November 2021 |
| G9R | Other | 30 | S30L | 1 November 2021 |
| J2L | Secretory | 54 | S54F | 1 November 2021 |
| J2R | Secretory | 54 | S54F | 1 November 2021 |
| A47R | Secretory | 221 | H221Y | 1 January 2022 |
| B21R | Other | 722 | P722S | 1 January 2022 |
| C19L | Membrane | 353 | E353K | 1 January 2022 |
| G10R | Membrane | 142 | M142I | 1 January 2022 |
| G9R | Other | 88 | D88N | 1 January 2022 |
| M4R | Other | 162 | E162K | 1 January 2022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Shi, H.; Cheng, G. Mpox Virus: Its Molecular Evolution and Potential Impact on Viral Epidemiology. Viruses 2023, 15, 995. https://doi.org/10.3390/v15040995
Yu X, Shi H, Cheng G. Mpox Virus: Its Molecular Evolution and Potential Impact on Viral Epidemiology. Viruses. 2023; 15(4):995. https://doi.org/10.3390/v15040995
Chicago/Turabian StyleYu, Xi, Huicheng Shi, and Gong Cheng. 2023. "Mpox Virus: Its Molecular Evolution and Potential Impact on Viral Epidemiology" Viruses 15, no. 4: 995. https://doi.org/10.3390/v15040995





