Phage-Host Prediction Using a Computational Tool Coupled with 16S rRNA Gene Amplicon Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Sample Collection
2.2. Bacterial DNA Extraction, Amplification and Sequencing
2.3. 16S rRNA Gene Sequence Analysis
2.4. Viral Particles Isolation and Viral Metagenomic Analysis
2.5. Identification and Classification of Phage Contigs
2.6. Prediction of Phage Hosts
2.7. Statistical Analysis
3. Results
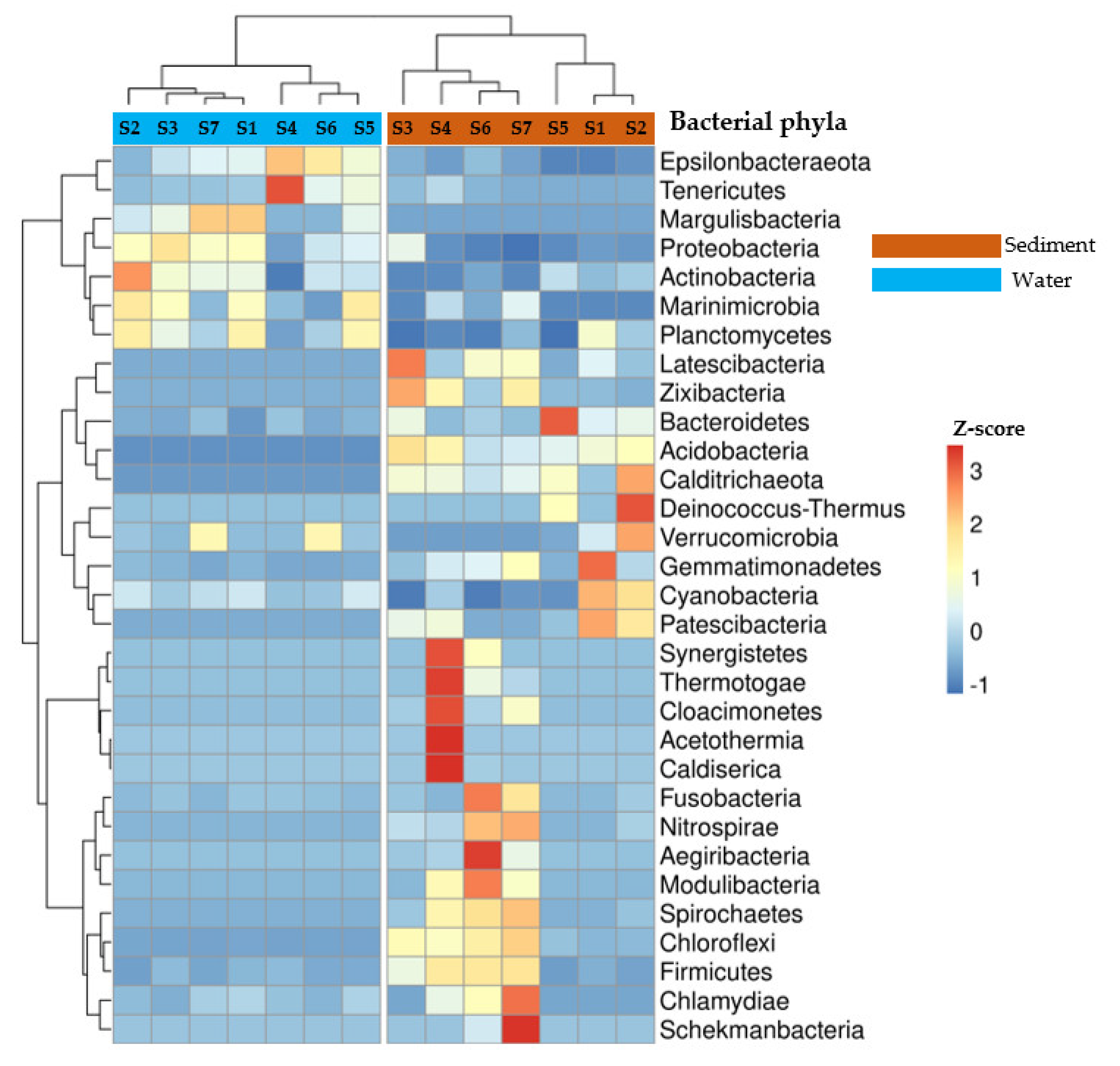
3.1. Taxonomic Profile of Bacterial Communities at Phyla Level in Water and Sediment Samples
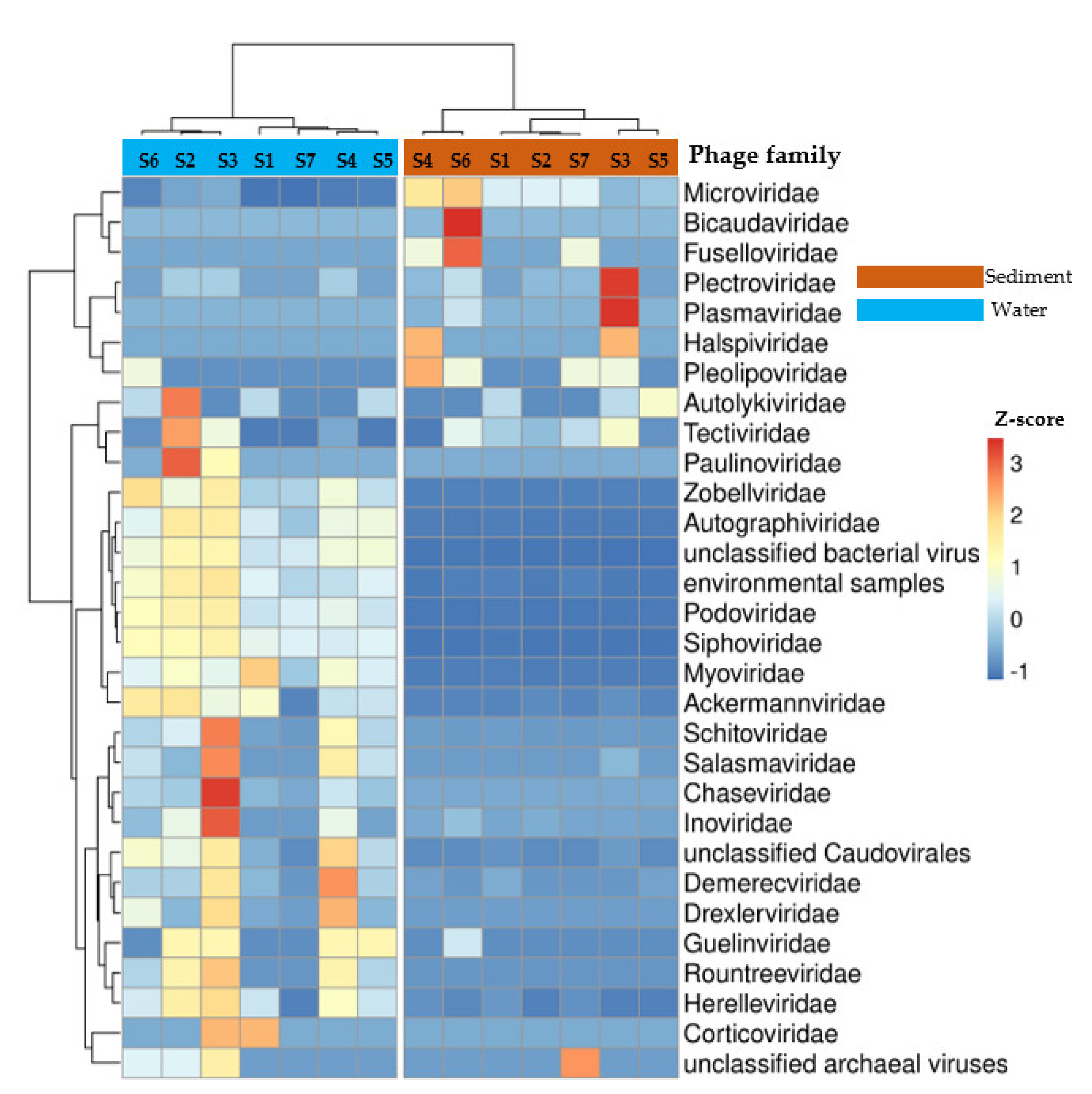
3.2. Taxonomic Profile of Phage Communities in Water and Sediment Samples
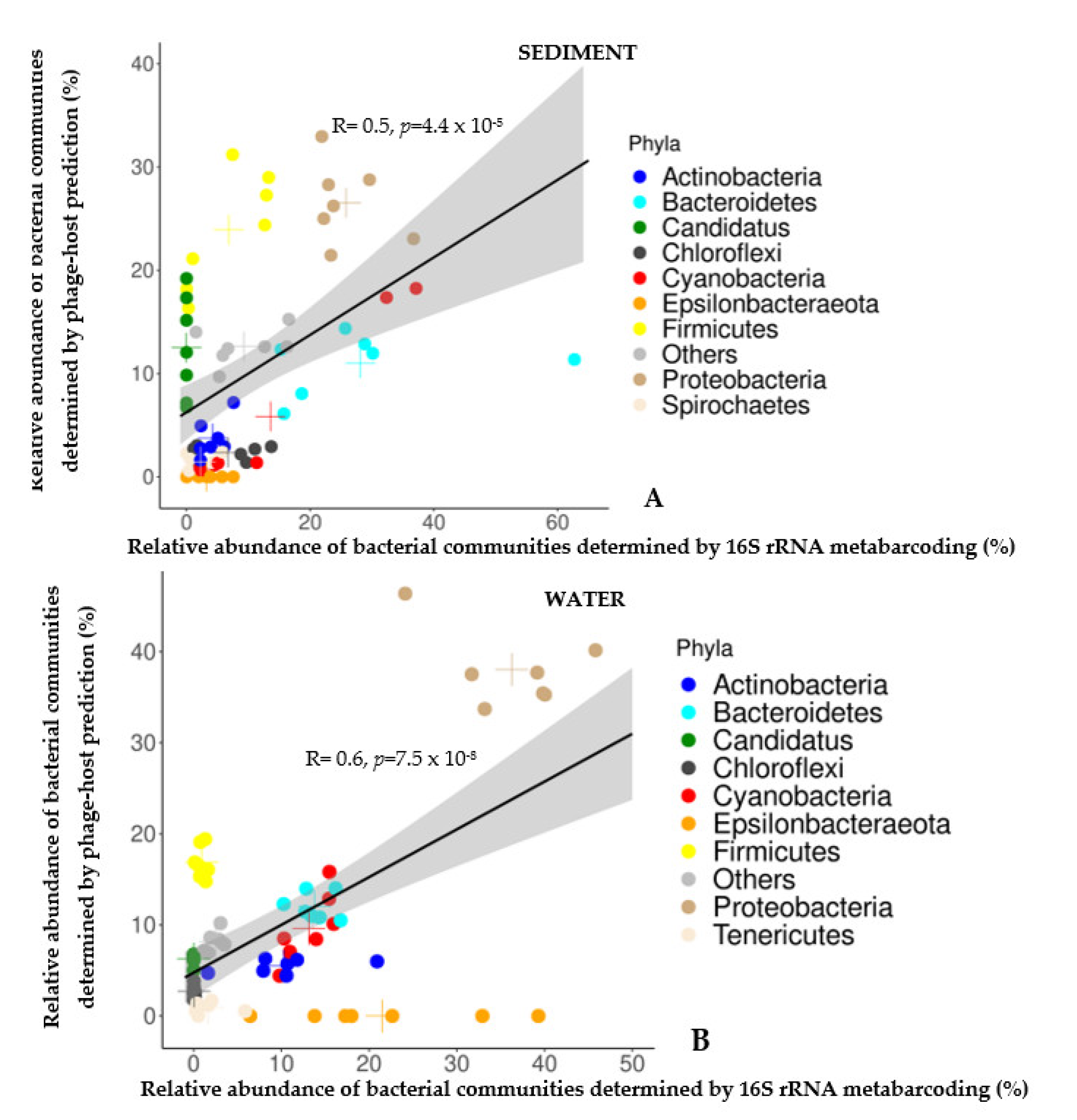
3.3. Comparison of the Relative Abundances of Bacterial Communities (At Major Phyla Level) Determined by Phage-Host Prediction (HP) and 16S Metabarcoding (16S)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guerin, E.; Hill, C. Shining Light on Human Gut Bacteriophages. Front. Cell. Infect. Microbiol. 2020, 10, 481. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Viruses in the Sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Clokie, M.R.; Millard, A.D.; Letarov, A.V.; Heaphy, S. Phages in Nature. Bacteriophage 2011, 1, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Bonnain, C.; Malki, K.; Sawaya, N.A. Phage Puppet Masters of the Marine Microbial Realm. Nat. Microbiol. 2018, 3, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, Z.; Abedon, S.T. Diversity of Phage Infection Types and Associated Terminology: The Problem with ‘Lytic or Lysogenic’. FEMS Microbiol. Lett. 2016, 363, fnw047. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, P.A.; Nobrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and Evolutionary Determinants of Bacteriophage Host Range. Trends Microbiol. 2019, 27, 51–63. [Google Scholar] [CrossRef]
- Moon, K.; Cho, J.-C. Metaviromics Coupled with Phage-Host Identification to Open the Viral ‘Black Box’. J. Microbiol. 2021, 59, 311–323. [Google Scholar] [CrossRef]
- Edwards, R.A.; McNair, K.; Faust, K.; Raes, J.; Dutilh, B.E. Computational Approaches to Predict Bacteriophage–Host Relationships. FEMS Microbiol. Rev. 2016, 40, 258–272. [Google Scholar] [CrossRef]
- Young, F.; Rogers, S.; Robertson, D.L. Predicting Host Taxonomic Information from Viral Genomes: A Comparison of Feature Representations. PLoS Comput. Biol. 2020, 16, e1007894. [Google Scholar] [CrossRef]
- Tan, J.; Fang, Z.; Wu, S.; Guo, Q.; Jiang, X.; Zhu, H. HoPhage: An Ab Initio Tool for Identifying Hosts of Phage Fragments from Metaviromes. Bioinformatics 2022, 38, 543–545. [Google Scholar] [CrossRef]
- Lu, C.; Zhang, Z.; Cai, Z.; Zhu, Z.; Qiu, Y.; Wu, A.; Jiang, T.; Zheng, H.; Peng, Y. Prokaryotic Virus Host Predictor: A Gaussian Model for Host Prediction of Prokaryotic Viruses in Metagenomics. BMC Biol. 2021, 19, 5. [Google Scholar] [CrossRef] [PubMed]
- Ahlgren, N.A.; Ren, J.; Lu, Y.Y.; Fuhrman, J.A.; Sun, F. Alignment-Free d∗2 Oligonucleotide Frequency Dissimilarity Measure Improves Prediction of Hosts from Metagenomically-Derived Viral Sequences. Nucleic Acids Res. 2017, 45, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Villarroel, J.; Kleinheinz, K.A.; Jurtz, V.I.; Zschach, H.; Lund, O.; Nielsen, M.; Larsen, M.V. HostPhinder: A Phage Host Prediction Tool. Viruses 2016, 8, 116. [Google Scholar] [CrossRef]
- Galiez, C.; Siebert, M.; Enault, F.; Vincent, J.; Söding, J. WIsH: Who Is the Host? Predicting Prokaryotic Hosts from Metagenomic Phage Contigs. Bioinformatics 2017, 33, 3113–3114. [Google Scholar] [CrossRef]
- Coutinho, F.H.; Zaragoza-Solas, A.; López-Pérez, M.; Barylski, J.; Zielezinski, A.; Dutilh, B.E.; Edwards, R.; Rodriguez-Valera, F. RaFAH: Host Prediction for Viruses of Bacteria and Archaea Based on Protein Content. Patterns 2021, 2, 100274. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, T.; Zhou, Y.; Han, Y.; Xu, M.; Gu, J. AfterQC: Automatic Filtering, Trimming, Error Removing and Quality Control for Fastq Data. BMC Bioinform. 2017, 18, 80. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data—ScienceOpen. Available online: https://www.scienceopen.com/document?vid=de674375-ab83-4595-afa9-4c8aa9e4e736 (accessed on 11 May 2022).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN Analysis of Metagenomic Data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining Viral Signal from Microbial Genomic Data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated Recovery, Annotation and Curation of Microbial Viruses, and Evaluation of Viral Community Function from Genomic Sequences. Microbiome 2020, 8, 90. [Google Scholar] [CrossRef]
- Noguchi, H.; Park, J.; Takagi, T. MetaGene: Prokaryotic Gene Finding from Environmental Genome Shotgun Sequences. Nucleic Acids Res. 2006, 34, 5623–5630. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The Protein Families Database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Jurtz, V.I.; Villarroel, J.; Lund, O.; Larsen, M.V.; Nielsen, M. MetaPhinder—Identifying Bacteriophage Sequences in Metagenomic Data Sets. PLoS ONE 2016, 11, e0163111. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Liu, R.; Qi, R.; Wang, J.; Zhang, Y.; Liu, X.; Rossetti, S.; Tandoi, V.; Yang, M. Phage-Host Associations in a Full-Scale Activated Sludge Plant during Sludge Bulking. Appl. Microbiol. Biotechnol. 2017, 101, 6495–6504. [Google Scholar] [CrossRef] [PubMed]
- Barylski, J.; Enault, F.; Dutilh, B.E.; Schuller, M.B.P.; Edwards, R.A.; Gillis, A.; Klumpp, J.; Knezevic, P.; Krupovic, M.; Kuhn, J.H.; et al. Analysis of Spounaviruses as a Case Study for the Overdue Reclassification of Tailed Bacteriophages. Syst. Biol. 2020, 69, 110–123. [Google Scholar] [CrossRef]
- Coutinho, F.H.; Gregoracci, G.B.; Walter, J.M.; Thompson, C.C.; Thompson, F.L. Metagenomics Sheds Light on the Ecology of Marine Microbes and Their Viruses. Trends Microbiol. 2018, 26, 955–965. [Google Scholar] [CrossRef]
- Bruder, K.; Maiki, K.; Cooper, A.; Sible, E.; Shapiro, J.W.; Watkins, S.C.; Putonti, C. Freshwater Metaviromics and Bacteriophages: A Current Assessment of the State of the Art in Relation to Bioinformatic Challenges: Supplementary Issue: Bioinformatics Methods and Applications for Big Metagenomics Data. Evol. Bioinform. 2016, 12, EBO-S38549. [Google Scholar] [CrossRef]
- Ly, M.; Abeles, S.R.; Boehm, T.K.; Robles-Sikisaka, R.; Naidu, M.; Santiago-Rodriguez, T.; Pride, D.T. Altered Oral Viral Ecology in Association with Periodontal Disease. mBio 2014, 5, e01133-14. [Google Scholar] [CrossRef]
- Coclet, C.; Roux, S. Global Overview and Major Challenges of Host Prediction Methods for Uncultivated Phages. Curr. Opin. Virol. 2021, 49, 117–126. [Google Scholar] [CrossRef]
- Jo, J.-H.; Kennedy, E.A.; Kong, H.H. Bacterial 16S Ribosomal RNA Gene Sequencing in Cutaneous Research. J. Investig. Derm. 2016, 136, e23–e27. [Google Scholar] [CrossRef]
- Peterson, D.; Bonham, K.S.; Rowland, S.; Pattanayak, C.W.; RESONANCE Consortium; Klepac-Ceraj, V.; Deoni, S.C.L.; D’Sa, V.; Bruchhage, M.; Volpe, A.; et al. Comparative Analysis of 16S RRNA Gene and Metagenome Sequencing in Pediatric Gut Microbiomes. Front. Microbiol. 2021, 12, 1651. [Google Scholar] [CrossRef]
- Větrovský, T.; Baldrian, P. The Variability of the 16S RRNA Gene in Bacterial Genomes and Its Consequences for Bacterial Community Analyses. PLoS ONE 2013, 8, e57923. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrianjakarivony, H.F.; Bettarel, Y.; Armougom, F.; Desnues, C. Phage-Host Prediction Using a Computational Tool Coupled with 16S rRNA Gene Amplicon Sequencing. Viruses 2023, 15, 76. https://doi.org/10.3390/v15010076
Andrianjakarivony HF, Bettarel Y, Armougom F, Desnues C. Phage-Host Prediction Using a Computational Tool Coupled with 16S rRNA Gene Amplicon Sequencing. Viruses. 2023; 15(1):76. https://doi.org/10.3390/v15010076
Chicago/Turabian StyleAndrianjakarivony, Harilanto Felana, Yvan Bettarel, Fabrice Armougom, and Christelle Desnues. 2023. "Phage-Host Prediction Using a Computational Tool Coupled with 16S rRNA Gene Amplicon Sequencing" Viruses 15, no. 1: 76. https://doi.org/10.3390/v15010076
APA StyleAndrianjakarivony, H. F., Bettarel, Y., Armougom, F., & Desnues, C. (2023). Phage-Host Prediction Using a Computational Tool Coupled with 16S rRNA Gene Amplicon Sequencing. Viruses, 15(1), 76. https://doi.org/10.3390/v15010076




