HSP27 Attenuates cGAS-Mediated IFN-β Signaling through Ubiquitination of cGAS and Promotes PRV Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus Strain
2.2. Plasmids and Reagents
2.3. Transfection and RNA Interference
2.4. RNA Extraction and Quantitative Real-Time PCR
2.5. Co-Immunoprecipitation and Western Blotting
2.6. PRV Infection and Infectivity Assays
2.7. Inhibitor Treatment Assay
2.8. cGAS Polyubiquitination Assay
2.9. Statistical Analysis
3. Results
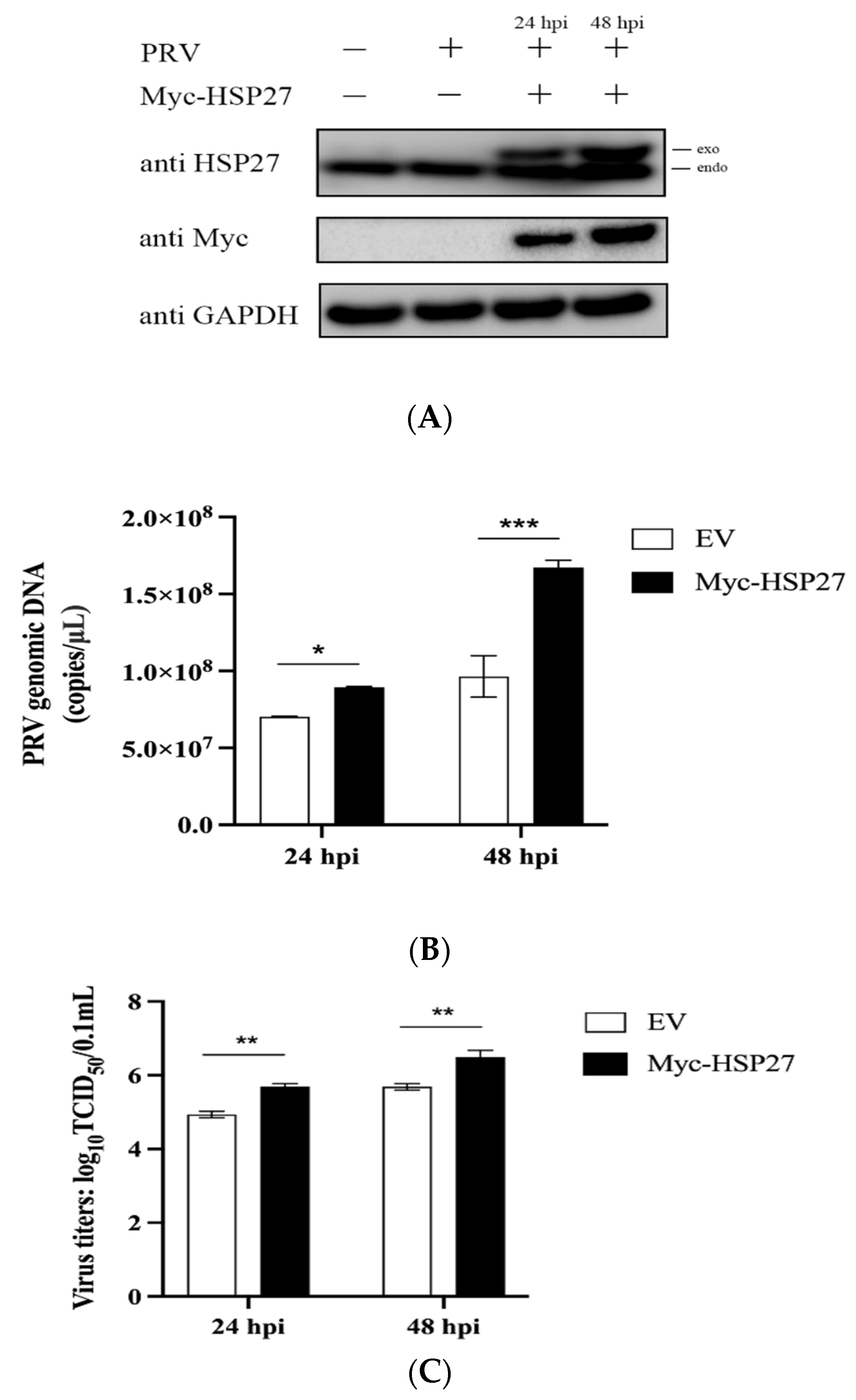
3.1. Overexpression of HSP27 Facilitates PRV Infection In Vitro
3.2. Interfering with HSP27 Restrains PRV Infection In Vitro
3.3. HSP27 Inhibits PRV Infection or Poly(dA:dT)-Activated IFN-β Expression
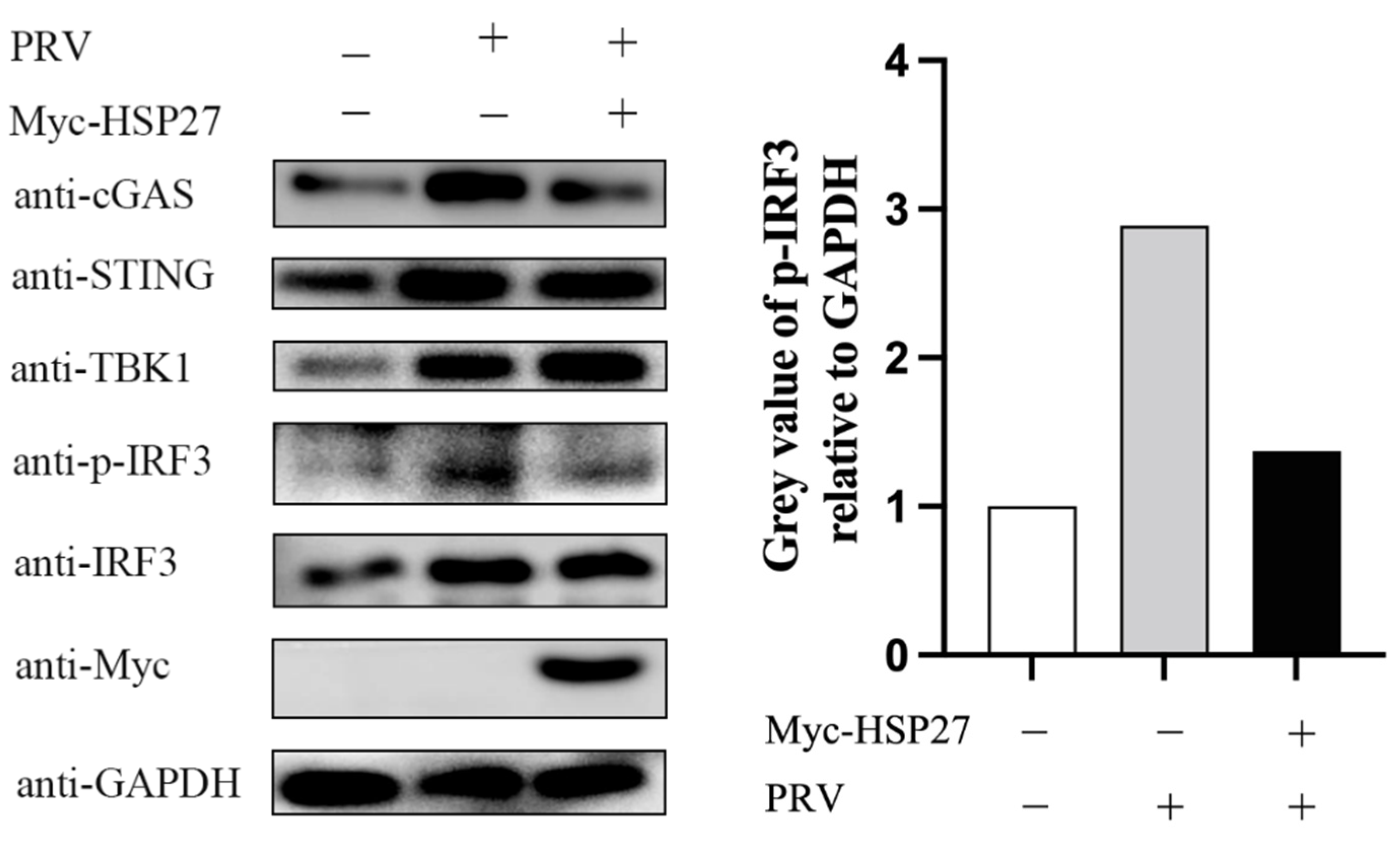
3.4. HSP27 Attenuates PRV-Triggered cGAS-Mediated Signaling Cascade
3.5. HSP27 Inhibits cGAS-STING-Mediated IFN-β Expression through Targeting cGAS
3.6. HSP27 Suppresses Endogenous cGAS Expression in Different Cells
3.7. HSP27 Interacts with cGAS and Promotes cGAS Ubiquitination
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, U.; Steinhoff, U.; Reis, L.F.; Hemmi, S.; Pavlovic, J.; Zinkernagel, R.M.; Aguet, M. Functional role of type I and type II interferons in antiviral defense. Science 1994, 264, 1918–1921. [Google Scholar] [CrossRef] [PubMed]
- Sadler, A.J.; Williams, B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Brennan, K.; Bowie, A. Activation of host pattern recognition receptors by viruses. Curr. Opin. Microbiol. 2010, 13, 503–507. [Google Scholar] [CrossRef]
- Xia, P.; Wang, S.; Gao, P.; Gao, G.; Fan, Z. DNA sensor cGAS-mediated immune recognition. Protein Cell 2016, 7, 777–791. [Google Scholar] [CrossRef] [Green Version]
- Wan, D.; Jiang, W.; Hao, J. Research Advances in How the cGAS-STING Pathway Controls the Cellular Inflammatory Response. Front. Immunol. 2020, 11, 615. [Google Scholar] [CrossRef]
- Koyama, S.; Ishii, K.; Coban, C.; Akira, S. Innate immune response to viral infection. Cytokine 2008, 43, 336–341. [Google Scholar] [CrossRef]
- Hu, M.M.; Shu, H.B. Innate Immune Response to Cytoplasmic DNA: Mechanisms and Diseases. Annu. Rev. Immunol. 2020, 38, 79–98. [Google Scholar] [CrossRef]
- Minamiguchi, K.; Kojima, S.; Sakumoto, K.; Kirisawa, R. Isolation and molecular characterization of a variant of Chinese gC-genotype II pseudorabies virus from a hunting dog infected by biting a wild boar in Japan and its pathogenicity in a mouse model. Virus Genes 2019, 55, 322–331. [Google Scholar] [CrossRef]
- Kong, H.; Zhang, K.; Liu, Y.; Shang, Y.; Wu, B.; Liu, X. Attenuated live vaccine (Bartha-K16) caused pseudorabies (Aujeszky’s disease) in sheep. Vet. Res. Commun. 2013, 37, 329–332. [Google Scholar] [CrossRef]
- Jin, H.L.; Gao, S.M.; Liu, Y.; Zhang, S.F.; Hu, R.L. Pseudorabies in farmed foxes fed pig offal in Shandong province, China. Arch. Virol. 2016, 161, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Kong, Z.; Liu, P.; Fu, Z.; Zhang, J.; Liu, M.; Shang, Y. Natural infection of a variant pseudorabies virus leads to bovine death in China. Transbound. Emerg. Dis. 2020, 67, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular biology of pseudorabies virus: Impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 2005, 69, 462–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Wang, M.; Cheng, A.; Yang, Q.; Wu, Y.; Jia, R.; Liu, M.; Zhu, D.; Chen, S.; Zhang, S. Innate Immune Evasion of Alphaherpesvirus Tegument Proteins. Front. Immunol. 2019, 10, 2196. [Google Scholar] [CrossRef] [Green Version]
- Lamote, J.A.S.; Kestens, M.; Van Waesberghe, C.; Delva, J.; De Pelsmaeker, S.; Devriendt, B.; Favoreel, H.W. The Pseudorabies Virus Glycoprotein gE/gI Complex Suppresses Type I Interferon Production by Plasmacytoid Dendritic Cells. J. Virol. 2017, 91, e02276-16. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Xu, A.; Qin, C.; Zhang, Q.; Chen, S.; Lang, Y.; Wang, M.; Li, C.; Feng, W.; Zhang, R.; et al. Pseudorabies Virus dUTPase UL50 Induces Lysosomal Degradation of Type I Interferon Receptor 1 and Antagonizes the Alpha Interferon Response. J. Virol. 2017, 91, e01148-17. [Google Scholar] [CrossRef] [Green Version]
- Qin, C.; Zhang, R.; Lang, Y.; Shao, A.; Xu, A.; Feng, W.; Han, J.; Wang, M.; He, W.; Yu, C.; et al. Bclaf1 critically regulates the type I interferon response and is degraded by alphaherpesvirus US3. PLoS Pathog. 2019, 15, e1007559. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Zhang, X.; Chen, L.; Bi, Y.; Idris, A.; Xu, S.; Li, X.; Zhang, Y.; Feng, R. Pseudorabies Virus US3 Protein Inhibits IFN-β Production by Interacting with IRF3 to Block Its Activation. Front. Microbiol. 2021, 12, 761282. [Google Scholar] [CrossRef]
- Lyu, C.; Li, W.D.; Wang, S.W.; Peng, J.M.; Yang, Y.B.; Tian, Z.J.; Cai, X.H. Host BAG3 Is Degraded by Pseudorabies Virus pUL56 C-Terminal 181L-185L and Plays a Negative Regulation Role during Viral Lytic Infection. Int. J. Mol. Sci. 2020, 21, 3148. [Google Scholar] [CrossRef]
- Lv, L.; Bai, J.; Gao, Y.; Jin, L.; Wang, X.; Cao, M.; Liu, X.; Jiang, P. Peroxiredoxin 1 Interacts with TBK1/IKKε and Negatively Regulates Pseudorabies Virus Propagation by Promoting Innate Immunity. J. Virol. 2021, 95, e0092321. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Bi, Y.; Xu, S.; Han, Y.; Idris, A.; Zhang, H.; Li, X.; Bai, J.; Zhang, Y.; Feng, R. Host antiviral protein IFITM2 restricts pseudorabies virus replication. Virus Res. 2020, 287, 198105. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, S.; Yang, X.; Wang, X.; Li, Y.; Wang, C.; Chen, L.; Chang, H. Porcine ISG15 modulates the antiviral response during pseudorabies virus replication. Gene 2018, 679, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, D.; Dong, S.; Zhai, H.; Kong, N.; Zheng, H.; Tong, W.; Li, G.; Shan, T.; Tong, G. Host Interferon-Stimulated Gene 20 Inhibits Pseudorabies Virus Proliferation. Virol. Sin. 2021, 36, 1027–1035. [Google Scholar] [CrossRef]
- Li, X.; Zhang, W.; Liu, Y.; Xie, J.; Hu, C.; Wang, X. Role of p53 in pseudorabies virus replication, pathogenicity, and host immune responses. Vet. Res. 2019, 50, 9. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Shi, H.; Qi, R.; Sun, S.; Tang, Y.; Zhang, B.; Wang, C. Hsp90 regulates activation of interferon regulatory factor 3 and TBK-1 stabilization in Sendai virus-infected cells. Mol. Biol. Cell 2006, 17, 1461–1471. [Google Scholar] [CrossRef] [Green Version]
- Acunzo, J.; Andrieu, C.; Baylot, V.; So, A.; Rocchi, P. Hsp27 as a therapeutic target in cancers. Curr. Drug Targets 2014, 15, 423–431. [Google Scholar] [CrossRef]
- Parcellier, A.; Schmitt, E.; Gurbuxani, S.; Seigneurin-Berny, D.; Pance, A.; Chantôme, A.; Plenchette, S.; Khochbin, S.; Solary, E.; Garrido, C. HSP27 is a ubiquitin-binding protein involved in I-kappaBalpha proteasomal degradation. Mol. Cell. Biol. 2003, 23, 5790–5802. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhang, L.; Zhu, X.; Bai, J.; Wang, L.; Wang, X.; Jiang, P. Heat shock protein 27 is involved in PCV2 infection in PK-15 cells. Virus Res. 2014, 189, 235–242. [Google Scholar] [CrossRef]
- Choi, Y.W.; Tan, Y.J.; Lim, S.G.; Hong, W.; Goh, P.Y. Proteomic approach identifies HSP27 as an interacting partner of the hepatitis C virus NS5A protein. Biochem. Biophys. Res. Commun. 2004, 318, 514–519. [Google Scholar] [CrossRef]
- Li, X.; Ma, R.; Wu, B.; Niu, Y.; Li, H.; Li, D.; Xie, J.; Idris, A.; Feng, R. HSP27 Protein Dampens Encephalomyocarditis Virus Replication by Stabilizing Melanoma Differentiation-Associated Gene 5. Front. Microbiol. 2021, 12, 788870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xie, J.; Gao, M.; Yan, Z.; Chen, L.; Wei, S.; Feng, R. Pseudorabies Virus ICP0 Abolishes Tumor Necrosis Factor Alpha-Induced NF-κB Activation by Degrading P65. Viruses 2022, 14, 954. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Xie, J.; Xu, S.; Bi, Y.; Li, X.; Zhang, H.; Idris, A.; Bai, J.; Feng, R. Encephalomyocarditis Virus Abrogates the Interferon Beta Signaling Pathway via Its Structural Protein VP2. J. Virol. 2021, 95, e01590-20. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, L. The interferon response to intracellular DNA: Why so many receptors? Immunobiology 2013, 218, 1312–1321. [Google Scholar] [CrossRef]
- Kong, Z.; Yin, H.; Wang, F.; Liu, Z.; Luan, X.; Sun, L.; Liu, W.; Shang, Y. Pseudorabies virus tegument protein UL13 recruits RNF5 to inhibit STING-mediated antiviral immunity. PLoS Pathog. 2022, 18, e1010544. [Google Scholar] [CrossRef]
- Zhang, Q.; Tang, Z.; An, R.; Ye, L.; Zhong, B. USP29 maintains the stability of cGAS and promotes cellular antiviral responses and autoimmunity. Cell Res. 2020, 30, 914–927. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.; Xie, C.; Ding, S.; Yang, H.; Guo, S.; Li, J.; Qin, L.; Ban, F.; Wang, D.; et al. A Novel Human Acute Encephalitis Caused by Pseudorabies Virus Variant Strain. Clin. Infect. Dis. 2021, 73, e3690–e3700. [Google Scholar] [CrossRef]
- Ran, Y.; Li, D.; Xiong, M.G.; Liu, H.N.; Feng, T.; Shi, Z.W.; Li, Y.H.; Wu, H.N.; Wang, S.Y.; Zheng, H.X. African swine fever virus I267L acts as an important virulence factor by inhibiting RNA polymerase III-RIG-I-mediated innate immunity. PLoS Pathog. 2022, 18, e1010270. [Google Scholar] [CrossRef]
- Sun, P.; Zhang, S.; Qin, X.; Chang, X.; Cui, X.; Li, H.; Zhang, S.; Gao, H.; Wang, P.; Zhang, Z.; et al. Foot-and-mouth disease virus capsid protein VP2 activates the cellular EIF2S1-ATF4 pathway and induces autophagy via HSPB1. Autophagy 2018, 14, 336–346. [Google Scholar] [CrossRef]
- Ling, S.; Luo, M.; Jiang, S.; Liu, J.; Ding, C.; Zhang, Q.; Guo, H.; Gong, W.; Tu, C.; Sun, J. Cellular Hsp27 interacts with classical swine fever virus NS5A protein and negatively regulates viral replication by the NF-κB signaling pathway. Virology 2018, 518, 202–209. [Google Scholar] [CrossRef]
- Yang, X.; Shi, C.; Li, H.; Shen, S.; Su, C.; Yin, H. MARCH8 attenuates cGAS-mediated innate immune responses through ubiquitylation. Sci. Signal. 2022, 15, eabk3067. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Huang, Y.J.; He, X.; Zhao, M.; Wang, X.; Liu, Z.S.; Xue, W.; Cai, H.; Zhan, X.Y.; Huang, S.Y.; et al. Acetylation Blocks cGAS Activity and Inhibits Self-DNA-Induced Autoimmunity. Cell 2019, 176, 1447–1460.e14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, C.; Yang, X.; Liu, Y.; Li, H.; Chu, H.; Li, G.; Yin, H. ZDHHC18 negatively regulates cGAS-mediated innate immunity through palmitoylation. EMBO J. 2022, 41, e109272. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Xie, J.; Li, D.; Li, H.; Niu, Y.; Wu, B.; Yang, Y.; Yan, Z.; Zhang, X.; Chen, L.; et al. HSP27 Attenuates cGAS-Mediated IFN-β Signaling through Ubiquitination of cGAS and Promotes PRV Infection. Viruses 2022, 14, 1851. https://doi.org/10.3390/v14091851
Li X, Xie J, Li D, Li H, Niu Y, Wu B, Yang Y, Yan Z, Zhang X, Chen L, et al. HSP27 Attenuates cGAS-Mediated IFN-β Signaling through Ubiquitination of cGAS and Promotes PRV Infection. Viruses. 2022; 14(9):1851. https://doi.org/10.3390/v14091851
Chicago/Turabian StyleLi, Xiangrong, Jingying Xie, Dianyu Li, Hongshan Li, Yuhui Niu, Bei Wu, Yanmei Yang, Zhenfang Yan, Xiangbo Zhang, Lei Chen, and et al. 2022. "HSP27 Attenuates cGAS-Mediated IFN-β Signaling through Ubiquitination of cGAS and Promotes PRV Infection" Viruses 14, no. 9: 1851. https://doi.org/10.3390/v14091851





