New Emergence of the Novel Pestivirus Linda Virus in a Pig Farm in Carinthia, Austria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Farm Description
2.2. Diagnostic Samples
2.3. Pathological Examination and Immunohistochemistry
2.4. Peripheral Blood Mononuclear Cell (PBMC) Isolation
2.5. Cell Culture
2.6. Indirect Immunofluorescence Assay
2.7. Virus Isolation
2.8. Serum Virus Neutralization (SVN) Assay
2.9. RNA Extraction and LindaV-Specific RT-qPCR Assay
2.10. RT-PCRs, Sanger Sequencing and Sequence Analysis
2.11. Phylogenetic Analysis
3. Results
3.1. Description of the Novel LindaV Outbreak
3.2. Diagnostic Workup
3.2.1. Serum Virus Neutralization Assay
3.2.2. LindaV-Specific RT-qPCR Assay
3.2.3. Virus Isolation
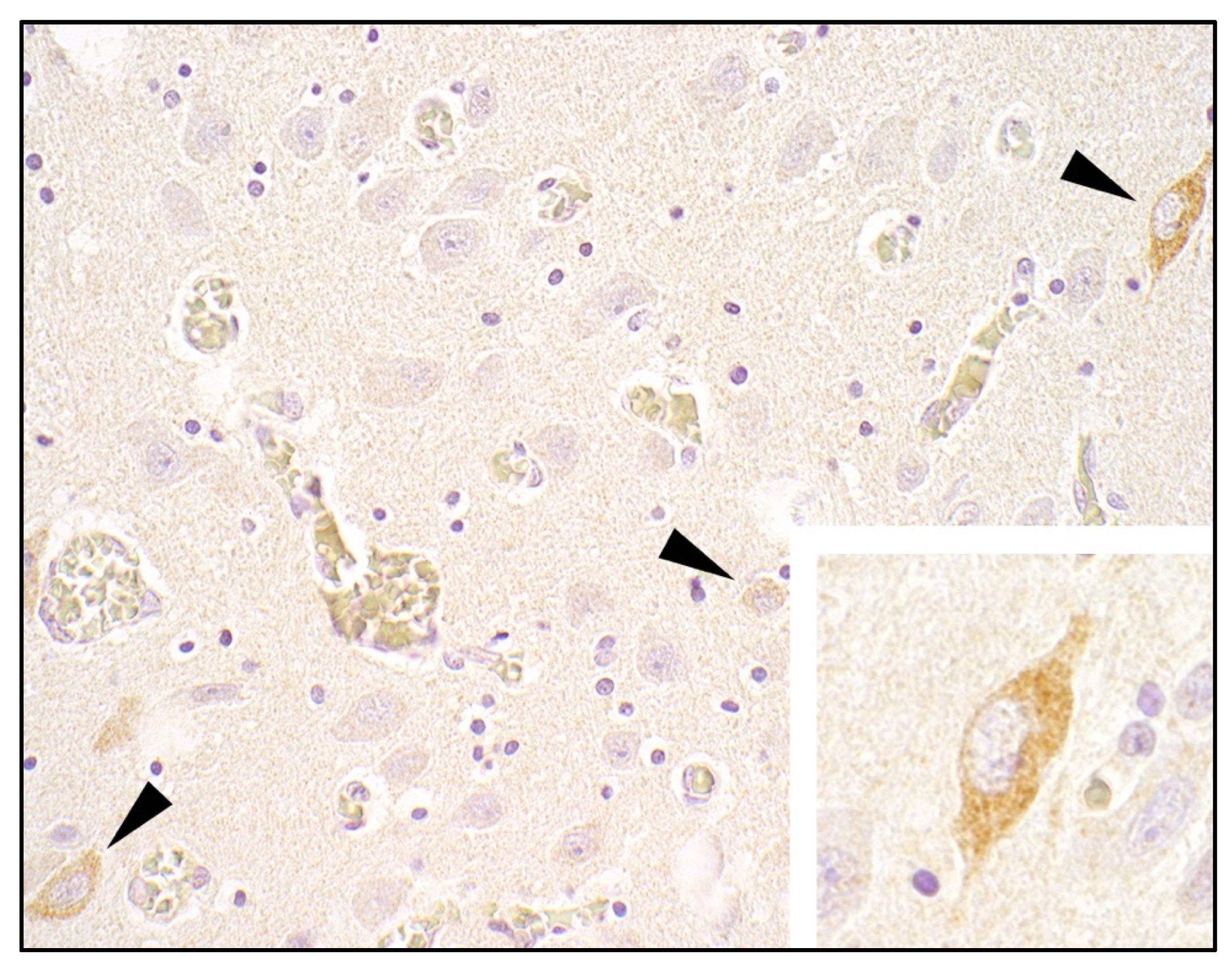
3.2.4. Histopathology and Immunohistochemistry
3.3. Genetic Characterization of the Novel LindaV Strain Austria3
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- OIE. Terrestrial Animal Health Code. Available online: https://www.oie.int/en/what-we-do/standards/codes-and-manuals/terrestrial-code-online-access/ (accessed on 9 October 2021).
- Kirkland, P.D.; Frost, M.J.; Finlaison, D.S.; King, K.R.; Ridpath, J.F.; Gu, X. Identification of a novel virus in pigs-bungowannah virus: A possible new species of pestivirus. Virus Res. 2007, 129, 26–34. [Google Scholar] [CrossRef]
- Hause, B.M.; Collin, E.A.; Peddireddi, L.; Yuan, F.; Chen, Z.; Hesse, R.A.; Gauger, P.C.; Clement, T.; Fang, Y.; Anderson, G. Discovery of a novel putative atypical porcine pestivirus in pigs in the USA. J. Gen. Virol. 2015, 96, 2994–2998. [Google Scholar] [CrossRef]
- Lamp, B.; Schwarz, L.; Högler, S.; Riedel, C.; Sinn, L.; Rebel-Bauder, B.; Weissenböck, H.; Ladinig, A.; Rümenapf, T. Novel pestivirus species in pigs, Austria, 2015. Emerg. Infect. Dis. 2017, 23, 1176–1179. [Google Scholar] [CrossRef]
- Kirkland, P.D.; Read, A.J.; Frost, M.J.; Finlaison, D.S. Bungowannah virus—A probable new species of pestivirus—What have we found in the last 10 years? Anim. Health Res. Rev. 2015, 16, 60–63. [Google Scholar] [CrossRef]
- Michelitsch, A.; Dalmann, A.; Wernike, K.; Reimann, I.; Beer, M. Seroprevalences of newly discovered porcine pestiviruses in german pig farms. Vet. Sci. 2019, 6, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abrahante, J.E.; Zhang, J.W.; Rossow, K.; Zimmerman, J.J.; Murtaugh, M.P. Surveillance of bungowannah pestivirus in the upper midwestern USA. Transbound. Emerg. Dis. 2014, 61, 375–377. [Google Scholar] [CrossRef]
- Cagatay, G.N.; Antos, A.; Meyer, D.; Maistrelli, C.; Keuling, O.; Becher, P.; Postel, A. Frequent infection of wild boar with atypical porcine pestivirus (APPV). Transbound. Emerg. Dis. 2018, 65, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Mósena AC, S.; Weber, M.N.; Cibulski, S.P.; Silva, M.S.; Paim, W.P.; Silva, G.S.; Medeiros, A.A.; Viana, N.A.; Baumbach, L.F.; Puhl, D.E.; et al. Survey for pestiviruses in backyard pigs in southern Brazil. J. Vet. Diagn. Investig. 2020, 32, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Han, Z.; Li, J.; Huang, Y.; Yang, J.; Ding, H.; Zhang, J.; Zhu, M.; Zhang, Y.; Liao, J.; et al. Atypical porcine pestivirus as a novel type of pestivirus in pigs in China. Front. Microbiol. 2017, 8, 862. [Google Scholar] [CrossRef] [PubMed]
- Beer, M.; Wernike, K.; Dräger, C.; Höper, D.; Pohlmann, A.; Bergermann, C.; Schröder, C.; Klinkhammer, S.; Blome, S.; Hoffmann, B. High prevalence of highly variable atypical porcine pestiviruses found in Germany. Transbound. Emerg. Dis. 2017, 64, e22–e26. [Google Scholar] [CrossRef]
- Kaufmann, C.; Stalder, H.; Sidler, X.; Renzullo, S.; Gurtner, C.; Grahofer, A.; Schweizer, M. Long-term circulation of atypical porcine pestivirus (APPV) within Switzerland. Viruses 2019, 11, 653. [Google Scholar] [CrossRef] [Green Version]
- Postel, A.; Meyer, D.; Cagatay, G.N.; Feliziani, F.; De Mia, G.M.; Fischer, N.; Grundhoff, A.; Milićević, V.; Deng, M.C.; Chang, C.Y.; et al. High abundance and genetic variability of atypical porcine pestivirus in pigs from Europe and Asia. Emerg. Infect. Dis. 2017, 23, 2104–2107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, L.; Riedel, C.; Högler, S.; Sinn, L.J.; Voglmayr, T.; Wöchtl, B.; Dinhopl, N.; Rebel-Bauder, B.; Weissenböck, H.; Ladinig, A.; et al. Congenital infection with atypical porcine pestivirus (APPV) is associated with disease and viral persistence. Vet. Res. 2017, 48, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiesler, A.; Seitz, K.; Schwarz, L.; Buczolich, K.; Petznek, H.; Sassu, E.; Dürlinger, S.; Högler, S.; Klang, A.; Riedel, C.; et al. Clinical and serological evaluation of LINDA virus infections in post-weaning piglets. Viruses 2019, 11, 975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiesler, A.; Plankensteiner, J.; Schwarz, L.; Riedel, C.; Seitz, K.; Mötz, M.; Ladinig, A.; Lamp, B.; Rümenapf, T. Prevalence of linda virus neutralizing antibodies in the austrian pig population. Viruses 2021, 13, 1001. [Google Scholar] [CrossRef]
- Kasza, L.; Shadduck, J.A.; Christofinis, G.J. Establishment, viral susceptibility and biological characteristics of a swine kidney cell line SK-6. Res. Vet. Sci. 1972, 13, 46–51. [Google Scholar] [CrossRef]
- Hoffmann, B.; Depner, K.; Schirrmeier, H.; Beer, M. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods 2006, 136, 200–209. [Google Scholar] [CrossRef]
- Douglas, S.E. DNA strider. Mol. Biotechnol. 1995, 3, 37–45. [Google Scholar] [CrossRef]
- Marck, C. “DNA strider”: A “C” program for the fast analysis of DNA and protein sequences on the apple macintosh family of computers. Nucleic Acids Res. 1988, 16, 1829–1836. [Google Scholar] [CrossRef]
- McClurkin, A.W.; Littledike, E.T.; Cutlip, R.C.; Frank, G.H.; Coria, M.F.; Bolin, S.R. Production of cattle immunotolerant to bovine viral diarrhea virus. Can. J. Comp. Med. 1984, 48, 156–161. [Google Scholar]
- Meyer, H.; Liess, B.; Frey, H.-R.; Hermanns, W.; Trautwein, G. Experimental transplacental transmission of hog cholera virus in pigs. IV. Virological and serological studies in newborn piglets. Zentralbl. Vet. B 1981, 28, 659–668. [Google Scholar] [CrossRef]
- Hermanns, W.; Trautwein, G.; Meyer, H.; Liess, B. Experimental transplacental transmission of hog cholera virus in pigs. V. Immunopathological findings in newborn pigs. Zentralbl. Vet. B 1981, 28, 669–683. [Google Scholar] [CrossRef]
- Nettleton, P.F.; Gilmour, J.S.; Herring, J.A.; Sinclair, J.A. The production and survival of lambs persistently infected with border disease virus. Comp. Immunol. Microbiol. Infect. Dis. 1992, 15, 179–188. [Google Scholar] [CrossRef]
- Finlaison, D.S.; Kirkland, P.D. The outcome of porcine foetal infection with bungowannah virus is dependent on the stage of gestation at which infection occurs. Part 1: Serology and virology. Viruses 2020, 12, 691. [Google Scholar] [CrossRef] [PubMed]
- Fulton, R.W.; Briggs, R.E.; Payton, M.E.; Confer, A.W.; Saliki, J.T.; Ridpath, J.F.; Burge, L.J.; Duff, G.C. Maternally Derived humoral immunity to bovine viral diarrhea virus (BVDV) 1a, BVDV1b, BVDV2, bovine herpesvirus-1, parainfluenza-3 virus bovine respiratory syncytial virus, mannheimia haemolytica and pasteurella multocida in beef calves, antibody decline. Vaccine 2004, 22, 643–649. [Google Scholar] [CrossRef]
- Vandeputte, J.; Too, H.L.; Ng, F.K.; Chen, C.; Chai, K.K.; Liao, G.A. Adsorption of colostral antibodies against classical swine fever, persistence of maternal antibodies, and effect on response to vaccination in baby pigs. Am. J. Vet. Res. 2001, 62, 1805–1811. [Google Scholar] [CrossRef]
- Bielefeldt-Ohmann, H.; Ronsholt, L.; Bloch, I. Demonstration of bovine viral diarrhoea virus in peripheral blood mononuclear cells of persistently infected, clinically normal cattle. J. Gen. Virol. 1987, 68, 1971–1982. [Google Scholar] [CrossRef]
- Summerfield, A.; Hofmann, M.A.; McCullough, K.C. Low density blood granulocytic cells induced during classical swine fever are targets for virus infection. Vet. Immunol. Immunopathol. 1998, 63, 289–301. [Google Scholar] [CrossRef]
- Wu, Z.; Ren, X.; Yang, L.; Hu, Y.; Yang, J.; He, G.; Zhang, J.; Dong, J.; Sun, L.; Du, J.; et al. Virome analysis for identification of novel mammalian viruses in bat species from chinese provinces. J. Virol. 2012, 86, 10999–11012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firth, C.; Bhat, M.; Firth, M.A.; Williams, S.H.; Frye, M.J.; Simmonds, P.; Conte, J.M.; Ng, J.; Garcia, J.; Bhuva, N.P.; et al. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal rattus norvegicus in New York City. MBio 2014, 5, e01933-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.-H.; Lin, X.-D.; Chen, Y.-M.; Xie, C.-G.; Tan, Z.-Z.; Zhou, J.-J.; Chen, S.; Holmes, E.C.; Zhang, Y.-Z. Newly identified viral genomes in pangolins with fatal disease. Virus Evol. 2020, 6, veaa020. [Google Scholar] [CrossRef] [PubMed]
- Jo, W.K.; van Elk, C.; van de Bildt, M.; van Run, P.; Petry, M.; Jesse, S.T.; Jung, K.; Ludlow, M.; Kuiken, T.; Osterhaus, A. An evolutionary divergent pestivirus lacking the Npro gene systemically infects a whale species. Emerg. Microbes Infect. 2019, 8, 1383–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiesler, A.; Schwarz, L.; Riedel, C.; Högler, S.; Brunthaler, R.; Dimmel, K.; Auer, A.; Zaruba, M.; Mötz, M.; Seitz, K.; et al. New Emergence of the Novel Pestivirus Linda Virus in a Pig Farm in Carinthia, Austria. Viruses 2022, 14, 326. https://doi.org/10.3390/v14020326
Kiesler A, Schwarz L, Riedel C, Högler S, Brunthaler R, Dimmel K, Auer A, Zaruba M, Mötz M, Seitz K, et al. New Emergence of the Novel Pestivirus Linda Virus in a Pig Farm in Carinthia, Austria. Viruses. 2022; 14(2):326. https://doi.org/10.3390/v14020326
Chicago/Turabian StyleKiesler, Alexandra, Lukas Schwarz, Christiane Riedel, Sandra Högler, René Brunthaler, Katharina Dimmel, Angelika Auer, Marianne Zaruba, Marlene Mötz, Kerstin Seitz, and et al. 2022. "New Emergence of the Novel Pestivirus Linda Virus in a Pig Farm in Carinthia, Austria" Viruses 14, no. 2: 326. https://doi.org/10.3390/v14020326






