A Vaccine Strategy Based on the Identification of an Annular Ganglioside Binding Motif in Monkeypox Virus Protein E8L
Abstract
:1. Introduction
2. Materials and Methods
2.1. Structure of the E8L Protein
2.2. Docking of E8L on Lipid Raft Gangliosides
2.3. Prediction of Linear B Epitopes
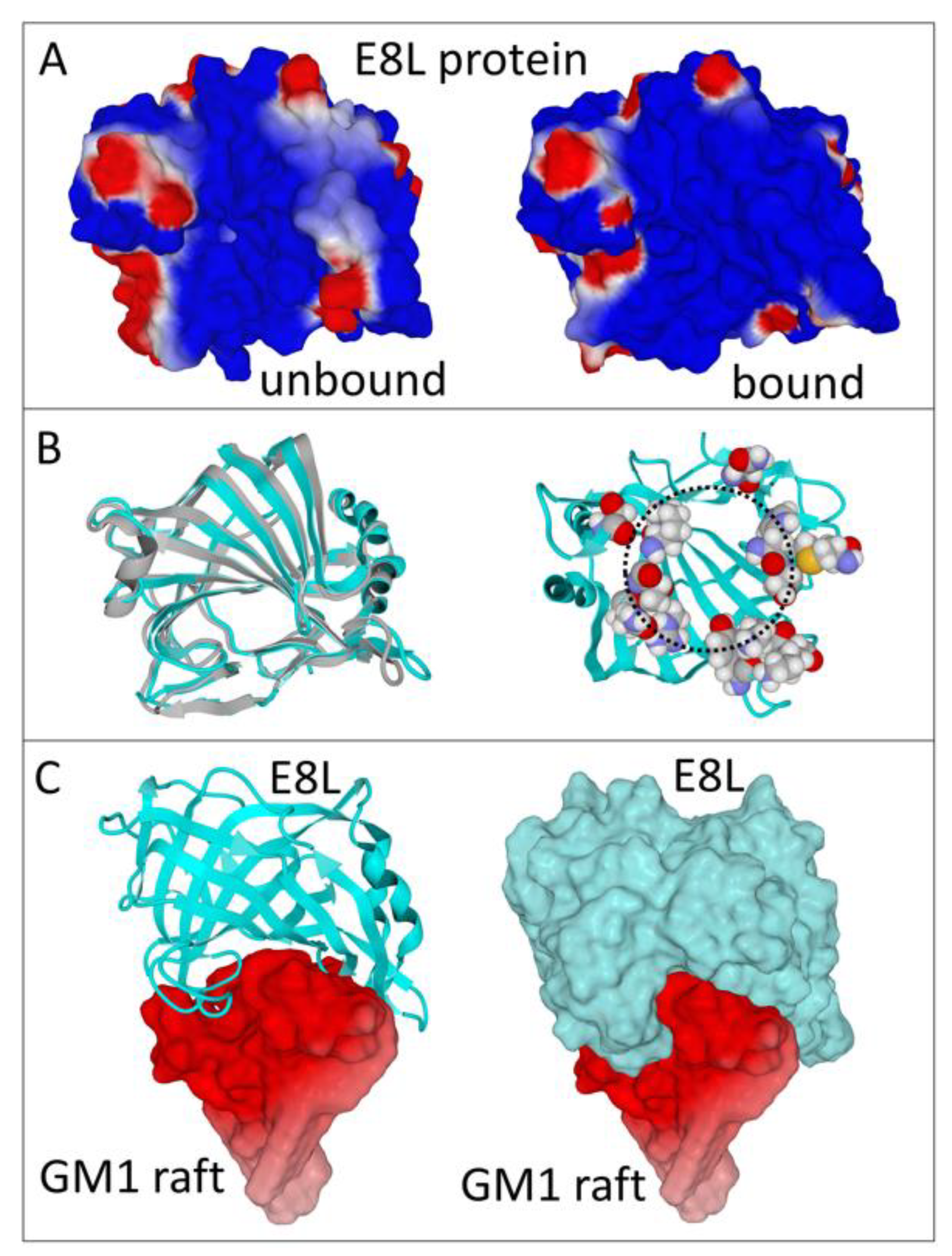
3. Results
3.1. Structure of E8L in the Unbound and Ganglioside-Bound Configurations
3.2. Identification of Surface-Exposed Linear B Epitopes in E8L
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Focosi, D.; Novazzi, F.; Baj, A.; Maggi, F. Monkeypox: An international epidemic. Rev. Med. Virol. 2022, 32, e2392. [Google Scholar] [CrossRef]
- CDC. Monkeypox: Vaccine Considerations. 2022; Updated 28 September 2022. Available online: https://www.cdc.gov/poxvirus/monkeypox/clinicians/vaccines/vaccine-considerations.html (accessed on 10 October 2022).
- Rao, A.K.; Petersen, B.W.; Whitehill, F.; Razeq, J.H.; Isaacs, S.N.; Merchlinsky, M.J.; Campos-Outcalt, D.; Morgan, R.L.; Damon, I.; Sánchez, P.J.; et al. Use of JYNNEOS (Smallpox and Monkeypox Vaccine, Live, Nonreplicating) for Preexposure Vaccination of Persons at Risk for Occupational Exposure to Orthopoxviruses: Recommendations of the Advisory Committee on Immunization Practices—United States. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, R.N.; Kennedy, J.S. ACAM2000: A newly licensed cell culture-based live vaccinia smallpox vaccine. Expert Opin. Investig. Drugs 2008, 17, 555–564. [Google Scholar] [CrossRef]
- See, K.C. Vaccination for Monkeypox Virus Infection in Humans: A Review of Key Considerations. Vaccines 2022, 10, 1342. [Google Scholar] [CrossRef] [PubMed]
- Alkhalil, A.; Strand, S.; Mucker, E.; Huggins, J.W.; Jahrling, P.B.; Ibrahim, S.M. Inhibition of monkeypox virus replication by RNA interference. Virol. J. 2009, 6, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, J.C.; Chung, C.S.; Chang, W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 1999, 73, 8750–8761. [Google Scholar] [CrossRef] [Green Version]
- Fantini, J.; Yahi, N.; Azzaz, F.; Chahinian, H. Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating COVID-19 outbreaks. J. Infect. 2021, 83, 197–206. [Google Scholar] [CrossRef]
- Azzaz, F.; Yahi, N.; Di Scala, C.; Chahinian, H.; Fantini, J. Ganglioside binding domains in proteins: Physiological and pathological mechanisms. Adv. Protein Chem. Struct. Biol. 2022, 128, 289–324. [Google Scholar]
- Fantini, J.; Chahinian, H.; Yahi, N. Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19. Biochem. Biophys. Res. Commun. 2021, 538, 132–136. [Google Scholar] [CrossRef]
- Verma, D.K.; Gupta, D.; Lal, S.K. Host Lipid Rafts Play a Major Role in Binding and Endocytosis of Influenza A Virus. Viruses 2018, 10, 650. [Google Scholar] [CrossRef] [Green Version]
- Engin, A.B.; Engin, E.D.; Engin, A. Dual function of sialic acid in gastrointestinal SARS-CoV-2 infection. Environ. Toxicol. Pharmacol. 2020, 79, 103436. [Google Scholar] [CrossRef] [PubMed]
- Sorice, M.; Misasi, R.; Riitano, G.; Manganelli, V.; Martellucci, S.; Longo, A.; Garofalo, T.; Mattei, V. Targeting Lipid Rafts as a Strategy Against Coronavirus. Front. Cell Dev. Biol. 2020, 8, 618296. [Google Scholar] [CrossRef] [PubMed]
- Villar, E.; Barroso, I.M. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: A minireview. Glycoconj. J. 2006, 23, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; McCord, K.A.; Bui, D.T.; Bouwman, K.M.; Kitova, E.N.; Elaish, M.; Kumawat, D.; Daskhan, G.C.; Tomris, I.; Han, L.; et al. Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2. Nat. Chem. Biol. 2022, 18, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. Solid-Phase Binding Assay for Ganglioside Binding of Human Respiroviruses. Methods Mol. Biol. 2022, 2556, 179–186. [Google Scholar]
- Luo, Y.; Motamedi, N.; Magaldi, T.G.; Gee, G.V.; Atwood, W.J.; DiMaio, D. Interaction between Simian Virus 40 Major Capsid Protein VP1 and Cell Surface Ganglioside GM1 Triggers Vacuole Formation. mBio 2016, 7, e00297. [Google Scholar] [CrossRef] [Green Version]
- Laliberte, J.P.; Moss, B. Lipid membranes in poxvirus replication. Viruses 2010, 2, 972–986. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.S.; Huang, C.Y.; Chang, W. Vaccinia virus penetration requires cholesterol and results in specific viral envelope proteins associated with lipid rafts. J. Virol. 2005, 79, 1623–1634. [Google Scholar] [CrossRef] [Green Version]
- Byrd, D.; Amet, T.; Hu, N.; Lan, J.; Hu, S.; Yu, Q. Primary human leukocyte subsets differentially express vaccinia virus receptors enriched in lipid rafts. J. Virol. 2013, 87, 9301–9312. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Li, D.; Duan, Z. Structural Basis of Glycan Recognition of Rotavirus. Front. Mol. Biosci. 2021, 8, 658029. [Google Scholar] [CrossRef]
- Delézay, O.; Hammache, D.; Fantini, J.; Yahi, N. SPC3, a V3 loop-derived synthetic peptide inhibitor of HIV-1 infection, binds to cell surface glycosphingolipids. Biochemistry 1996, 35, 15663–15671. [Google Scholar] [CrossRef] [PubMed]
- Hammache, D.; Yahi, N.; Piéroni, G.; Ariasi, F.; Tamalet, C.; Fantini, J. Sequential interaction of CD4 and HIV-1 gp120 with a reconstituted membrane patch of ganglioside GM3: Implications for the role of glycolipids as potential HIV-1 fusion cofactors. Biochem. Biophys. Res. Commun. 1998, 246, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Jaafar, R.; Boschi, C.; Aherfi, S.; Bancod, A.; Le Bideau, M.; Edouard, S.; Colson, P.; Chahinian, H.; Raoult, D.; Yahi, N.; et al. High Individual Heterogeneity of Neutralizing Activities against the Original Strain and Nine Different Variants of SARS-CoV-2. Viruses 2021, 13, 2177. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Fantini, J.; Yahi, N.; Delerce, J.; Levasseur, A.; Fournier, P.E.; Lagier, J.C.; Raoult, D.; La Scola, B. Limited spread of a rare spike E484K-harboring SARS-CoV-2 in Marseille. France. Arch. Virol. 2022, 167, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Di Scala, C.; Chahinian, H.; Yahi, N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents 2020, 55, 105960. [Google Scholar] [CrossRef]
- Fantini, J.; Chahinian, H.; Yahi, N. Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: What molecular dynamics studies of virus-host interactions reveal. Int. J. Antimicrob. Agents 2020, 56, 106020. [Google Scholar] [CrossRef]
- Azzaz, F.; Fantini, J. The epigenetic dimension of protein structure. Biomol. Concepts 2022, 13, 55–60. [Google Scholar] [CrossRef]
- Azzaz, F.; Yahi, N.; Chahinian, H.; Fantini, J. The Epigenetic Dimension of Protein Structure Is an Intrinsic Weakness of the AlphaFold Program. Biomolecules 2022, 12, 1527. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Colson, P.; Chahinian, H.; La Scola, B.; Raoult, D. The puzzling mutational landscape of the SARS-2-variant Omicron. J. Med. Virol. 2022, 94, 2019–2025. [Google Scholar] [CrossRef]
- Yahi, N.; Chahinian, H.; Fantini, J. Infection-enhancing anti-SARS-CoV-2 antibodies recognize both the original Wuhan/D614G strain and Delta variants. A potential risk for mass vaccination? J. Infect. 2021, 83, 607–635. [Google Scholar] [CrossRef]
- Colson, P.; Delerce, J.; Beye, M.; Levasseur, A.; Boschi, C.; Houhamdi, L.; Tissot-Dupont, H.; Yahi, N.; Million, M.; La Scola, B.; et al. First cases of infection with the 21L/BA.2 Omicron variant in Marseille. France. J. Med. Virol. 2022, 94, 3421–3430. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Fournier, P.E.; Delerce, J.; Million, M.; Bedotto, M.; Houhamdi, L.; Yahi, N.; Bayette, J.; Levasseur, A.; Fantini, J.; et al. Culture and identification of a "Deltamicron" SARS-CoV-2 in a three cases cluster in southern France. J. Med. Virol. 2022, 94, 3739–3749. [Google Scholar] [CrossRef] [PubMed]
- Guérin, P.; Yahi, N.; Azzaz, F.; Chahinian, H.; Sabatier, J.M.; Fantini, J. Structural Dynamics of the SARS-CoV-2 Spike Protein: A 2-Year Retrospective Analysis of SARS-CoV-2 Variants (from Alpha to Omicron) Reveals an Early Divergence between Conserved and Variable Epitopes. Molecules 2022, 27, 3851. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.L. The role of cell surface sialic acids for SARS-CoV-2 infection. Glycobiology 2021, 31, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V.; Nelli, R.K.; Gontu, A.; Satyakumar, R.; Surendran Nair, M.; Subbiah, M. Sialic Acid Receptors: The Key to Solving the Enigma of Zoonotic Virus Spillover. Viruses 2021, 13, 262. [Google Scholar] [CrossRef]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef]
- Parker, J.M.; Guo, D.; Hodges, R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: Correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 1986, 25, 5425–5432. [Google Scholar] [CrossRef]
- Karplus, P.; Schulz, G. Prediction of chain flexibility in proteins. Naturwissenschaften 1985, 72, 212–213. [Google Scholar] [CrossRef]
- Emini, E.A.; Hughes, J.V.; Perlow, D.; Boger, J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985, 55, 836–839. [Google Scholar] [CrossRef] [Green Version]
- Pellequer, J.-L.; Westhof, E.; Van Regenmortel, M.H. Correlation between the location of antigenic sites and the prediction of turns in proteins. Immunol. Lett. 1993, 36, 83–99. [Google Scholar] [CrossRef]
- Janin, J.; Wodak, S.; Levitt, M.; Maigret, B. Conformation of amino acid side-chains in proteins. J. Mol. Biol. 1978, 125, 357–386. [Google Scholar] [CrossRef]
- Ponnuswamy, P.; Prabhakaran, M.; Manavalan, P. Hydrophobic packing and spatial arrangement of amino acid residues in globular proteins. Biochim. Et Biophys. Acta (BBA)-Protein Struct. 1980, 623, 301–316. [Google Scholar] [CrossRef]
- Kolaskar, A.S.; Tongaonkar, P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990, 276, 172–174. [Google Scholar] [CrossRef] [Green Version]
- Clifford, J.; Hoeie, M.H.; Nielsen, M.; Deleuran, S.; Peters, B.; Marcatili, P. BepiPred-3.0: Improved B-cell epitope prediction using protein language models. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Raghava, G.P.S. BcePred: Prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. In International Conference on Artificial Immune Systems; Springer: Berlin/Heidelberg, Germany, 2004; pp. 197–204. [Google Scholar]
- Shantier, S.W.; Mustafa, M.I.; Abdelmoneim, A.H.; Fadl, H.A.; Elbager, S.G.; Makhawi, A.M. Novel multi epitope-based vaccine against monkeypox virus: Vaccinomic approach. Sci. Rep. 2022, 12, 15983. [Google Scholar] [CrossRef]
- El-Battari, A.; Rodriguez, L.; Chahinian, H.; Delézay, O.; Fantini, J.; Yahi, N.; Di Scala, C. Gene Therapy Strategy for Alzheimer’s and Parkinson’s Diseases Aimed at Preventing the Formation of Neurotoxic Oligomers in SH-SY5Y Cells. Int. J. Mol. Sci. 2021, 22, 11550. [Google Scholar] [CrossRef]
- Reimer, U. Prediction of linear B-cell epitopes. Methods Mol. Biol. 2009, 524, 335–344. [Google Scholar]
- Galanis, K.A.; Nastou, K.C.; Papandreou, N.C.; Petichakis, G.N.; Pigis, D.G.; Iconomidou, V.A. Linear B-Cell Epitope Prediction for In Silico Vaccine Design: A Performance Review of Methods Available via Command-Line Interface. Int. J. Mol. Sci. 2021, 22, 3210. [Google Scholar] [CrossRef]
- Kwon, S.; Kwon, M.; Im, S.; Lee, K.; Lee, H. mRNA vaccines: The most recent clinical applications of synthetic mRNA. Arch. Pharmacal Res. 2022, 45, 245–262. [Google Scholar] [CrossRef]
- Warren, C.J.; Yu, S.; Peters, D.K.; Barbachano-Guerrero, A.; Yang, Q.; Burris, B.L.; Worwa, G.; Huang, I.C.; Wilkerson, G.K.; Goldberg, T.L.; et al. Primate hemorrhagic fever-causing arteriviruses are poised for spillover to humans. Cell 2022, 185, 3980–3991.e18. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, Q.; Tang, J.; Feng, W.-H. Lipid rafts both in cellular membrane and viral envelope are critical for PRRSV efficient infection. Virology 2015, 484, 170–180. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fantini, J.; Chahinian, H.; Yahi, N. A Vaccine Strategy Based on the Identification of an Annular Ganglioside Binding Motif in Monkeypox Virus Protein E8L. Viruses 2022, 14, 2531. https://doi.org/10.3390/v14112531
Fantini J, Chahinian H, Yahi N. A Vaccine Strategy Based on the Identification of an Annular Ganglioside Binding Motif in Monkeypox Virus Protein E8L. Viruses. 2022; 14(11):2531. https://doi.org/10.3390/v14112531
Chicago/Turabian StyleFantini, Jacques, Henri Chahinian, and Nouara Yahi. 2022. "A Vaccine Strategy Based on the Identification of an Annular Ganglioside Binding Motif in Monkeypox Virus Protein E8L" Viruses 14, no. 11: 2531. https://doi.org/10.3390/v14112531




