Development of a Monoclonal Antibody Targeting HTLV-1 Envelope gp46 Glycoprotein and Its Application to Near-Infrared Photoimmuno-Antimicrobial Strategy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Plasmid Construction
2.3. Cell-Free Protein Synthesis and Purification
2.4. Immunization and Generation of Hybridomas
2.5. Purification of mAbs
2.6. Flow Cytometry Analysis
2.7. Immunofluorescence (IF)
2.8. Enzyme-Linked Immunosorbent Assay (ELISA)
2.9. AlphaScreen Analysis
2.10. Epitope Prediction
2.11. Affinity Measurement of mAbs
2.12. Reporter Cells Generation and Reporter Assay
2.13. HTLV-1 gp46 mAb Internalization Assay
2.14. Generation of IR700 Conjugated mAb
2.15. Cell Viability and Lactate Dehydrogenase (LDH) Assay
2.16. Live/Dead Cell Staining Assay
2.17. Reporter Assay after NIR-PIAS
2.18. Statistical Analysis
3. Results
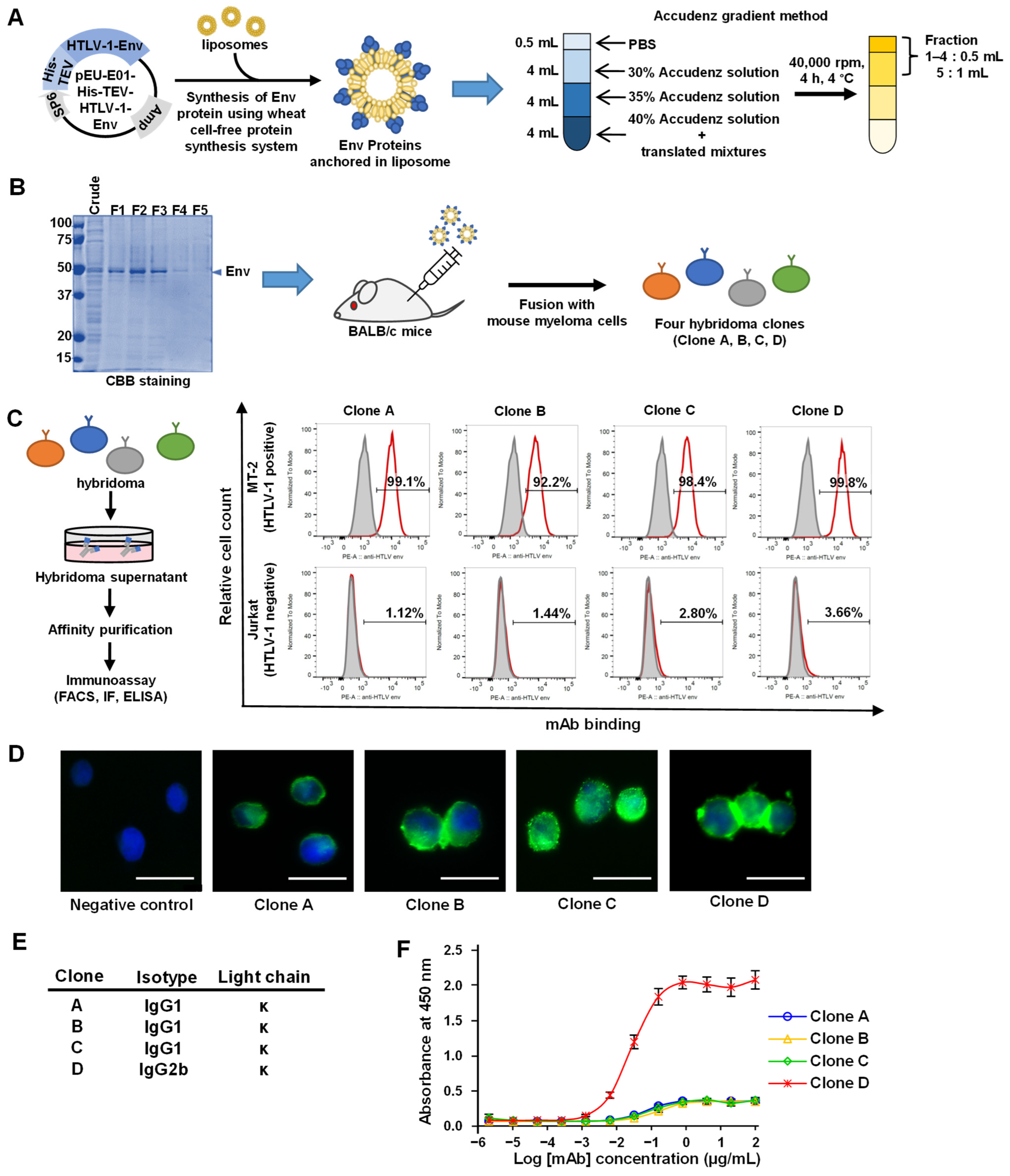
3.1. Synthesis of the HTLV-1 Env Protein and Production of mAbs
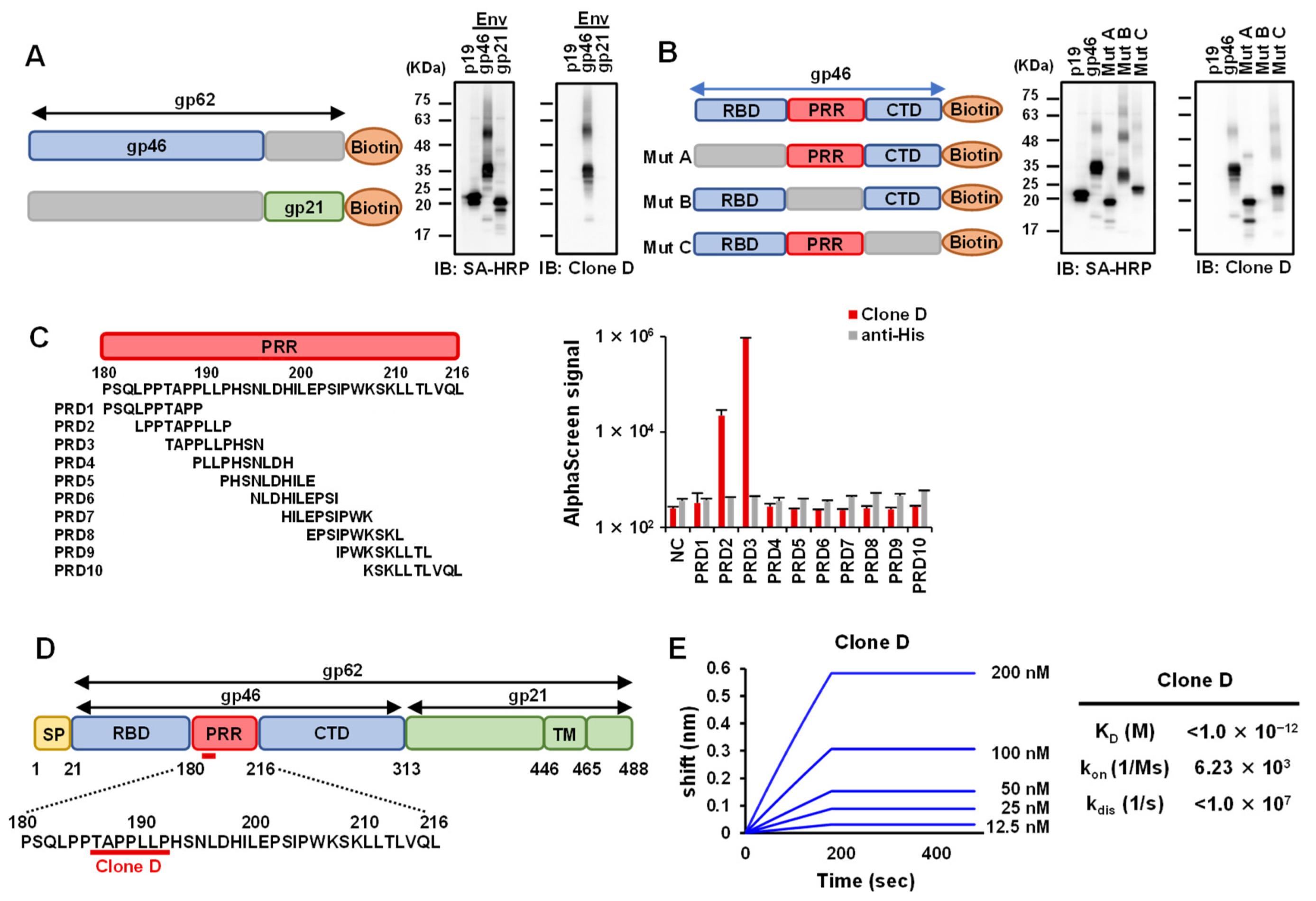
3.2. Epitope Analysis of Selected mAbs against HTLV-1 Env
3.3. Quantitative HTLV-1-Mediated Cell Fusion Assay Using secNL
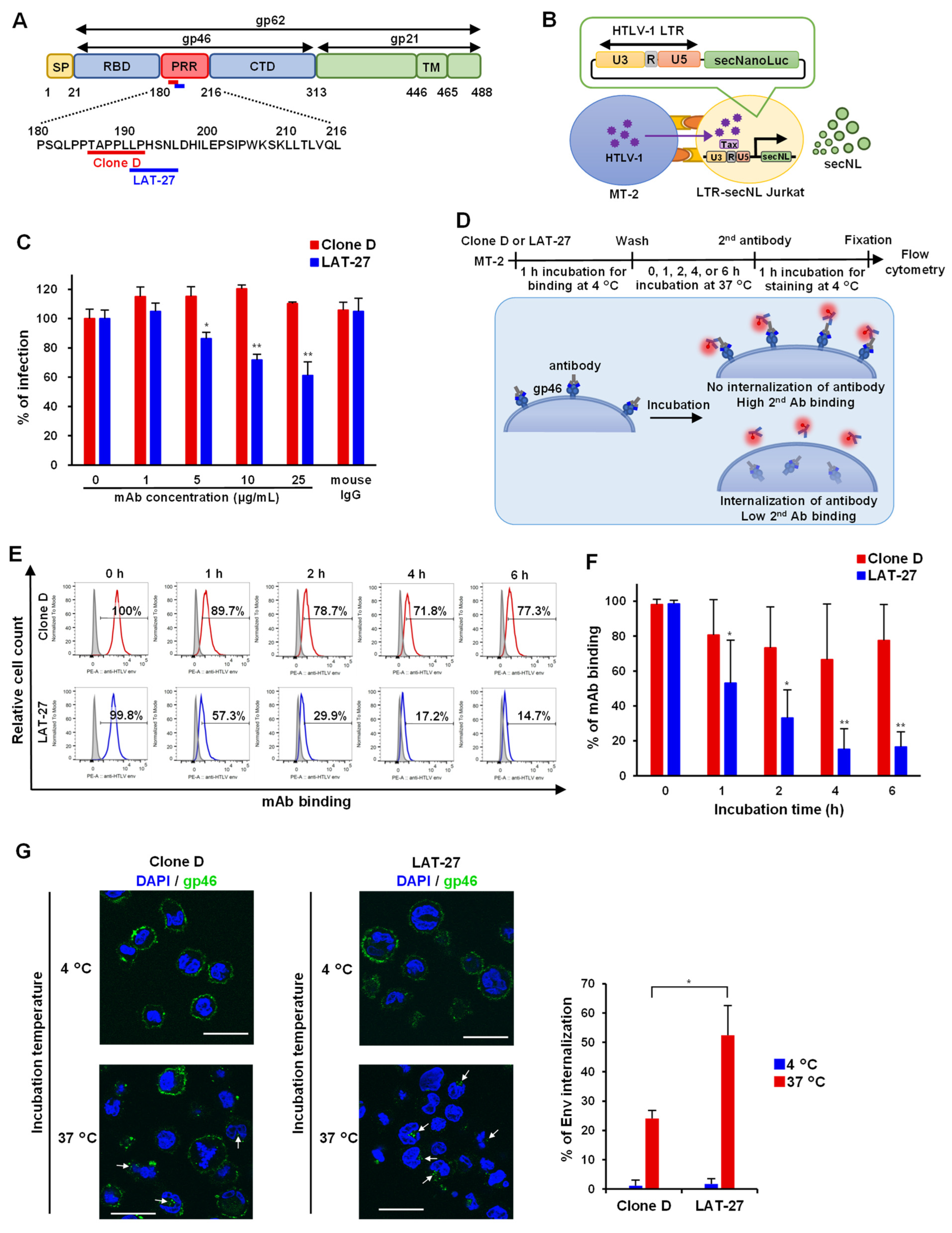
3.4. Internalization Assay of mAb against Anti-HTLV-1 Env
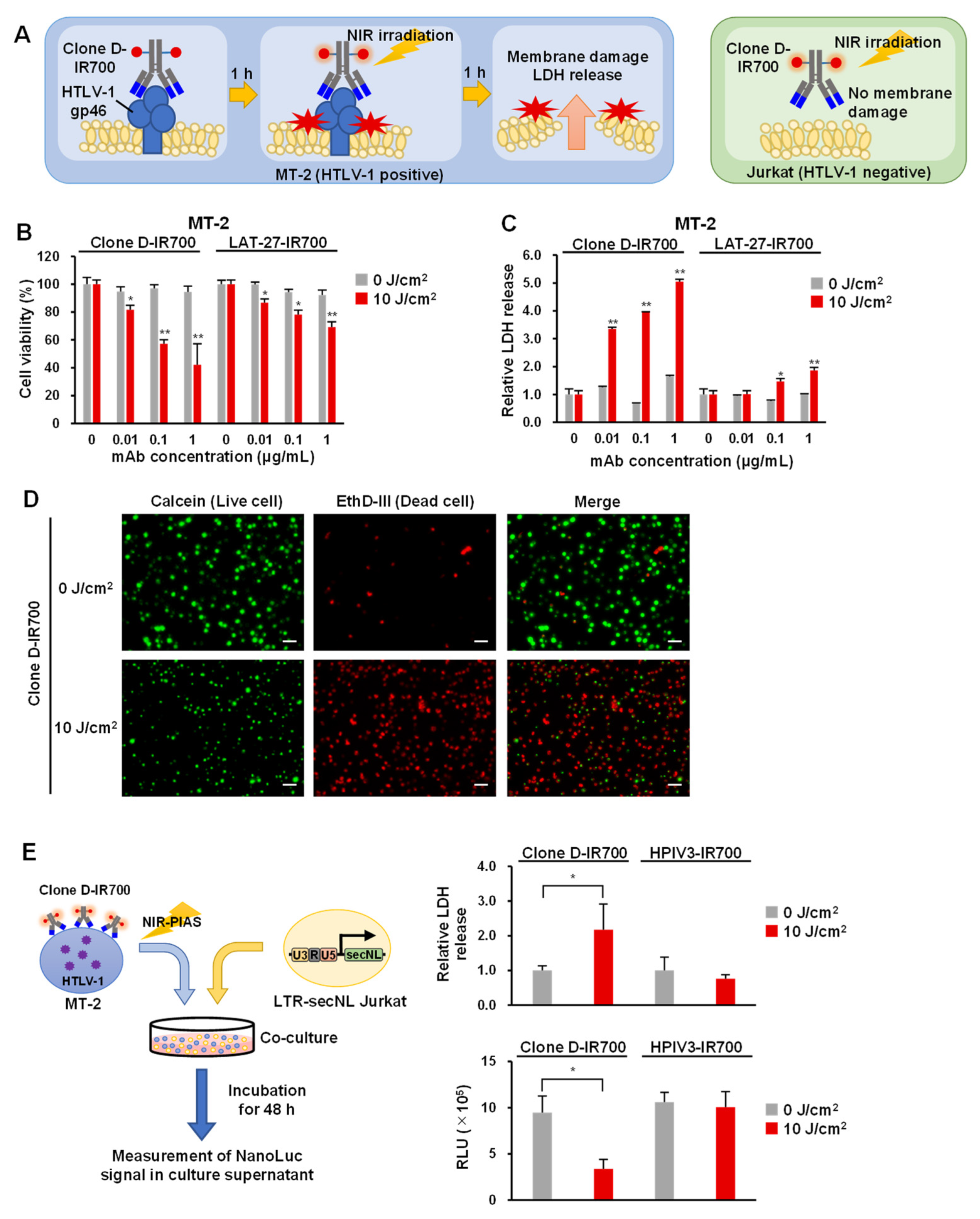
3.5. Application of Antibodies to NIR-PIAS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hinuma, Y.; Nagata, K.; Hanaoka, M.; Nakai, M.; Matsumoto, T.; Kinoshita, K.I.; Shirakawa, S.; Miyoshi, I. Adult T-cell leukemia: Antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. USA 1981, 78, 6476–6480. [Google Scholar] [CrossRef] [PubMed]
- Poiesz, B.J.; Ruscetti, F.W.; Gazdar, A.F.; Bunn, P.A.; Minna, J.D.; Gallo, R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 1980, 77, 7415–7419. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T. Human T cell leukemia virus type I (HTLV-I) and human diseases. Annu. Rev. Immunol. 1997, 15, 15–37. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Cassar, O. Epidemiological aspects and world distribution of HTLV-1 infection. Front. Microbiol. 2012, 3, 388. [Google Scholar] [CrossRef]
- Iwanaga, M.; Watanabe, T.; Utsunomiya, A.; Okayama, A.; Uchimaru, K.; Koh, K.-R.; Ogata, M.; Kikuchi, H.; Sagara, Y.; Uozumi, K.; et al. Human T-cell leukemia virus type I (HTLV-1) proviral load and disease progression in asymptomatic HTLV-1 carriers: A nationwide prospective study in Japan. Blood 2010, 116, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, E.; Ono, A.; Hikita, N.; Arima, K.; Mochizuki, M.; Yamaguchi, K.; Tajima, K.; Kiyokawa, H. Estimated Prevalence Rate of HTLV-I Uveitis in Chikugo. J. Jpn. Ophthalmol. Soc. 1998, 102, 327–332. [Google Scholar]
- Ohguro, N.; Sonoda, K.-H.; Takeuchi, M.; Matsumura, M.; Mochizuki, M. The 2009 prospective multi-center epidemiologic survey of uveitis in Japan. Jpn. J. Ophthalmol. 2012, 56, 432–435. [Google Scholar] [CrossRef]
- Saito, M. Neuroimmunological aspects of human T cell leukemia virus type 1-associated myelopathy/tropical spastic paraparesis. J. Neurovirol. 2014, 20, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, M. Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma: A REPORT FROM THE LYMPHOMA STUDY GROUP (1984–87). Br. J. Haematol. 1991, 79, 428–437. [Google Scholar] [CrossRef]
- Katsuya, H.; Ishitsuka, K.; Utsunomiya, A.; Hanada, S.; Eto, T.; Moriuchi, Y.; Saburi, Y.; Miyahara, M.; Sueoka, E.; Uike, N.; et al. Treatment and survival among 1594 patients with ATL on behalf of the ATL-Prognostic Index Project. Blood 2015, 126, 2570–2577. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Watanabe, T. Human T lymphotropic virus type-I and adult T-cell leukemia in Japan. Int. J. Hematol. 2002, 76 (Suppl. S2), 240–245. [Google Scholar] [CrossRef]
- Jones, K.S.; Lambert, S.; Bouttier, M.; Bénit, L.; Ruscetti, F.W.; Hermine, O.; Pique, C. Molecular aspects of HTLV-1 entry: Functional domains of the HTLV-1 surface subunit (SU) and their relationships to the entry receptors. Viruses 2011, 3, 794–810. [Google Scholar] [CrossRef]
- Ghez, D.; Lepelletier, Y.; Jones, K.S.; Pique, C.; Hermine, O. Current concepts regarding the HTLV-1 receptor complex. Retrovirology; BioMed Central: London, UK, 2010; Volume 7. [Google Scholar] [CrossRef]
- Gross, C.; Thoma-Kress, A.K. Molecular Mechanisms of HTLV-1 Cell-to-Cell Transmission. Viruses 2016, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.L.; Maldonado, J.O.; Mueller, J.D.; Zhang, W.; Mansky, L.M. Molecular Studies of HTLV-1 Replication: An Update. Viruses 2016, 8, 31. [Google Scholar] [CrossRef]
- Percher, F.; Jeannin, P.; Martin-Latil, S.; Gessain, A.; Afonso, P.V.; Vidy-Roche, A.; Ceccaldi, P.-E. Mother-to-Child Transmission of HTLV-1 Epidemiological Aspects, Mechanisms and Determinants of Mother-to-Child Transmission. Viruses 2016, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Hino, S.; Sugiyama, H.; Doi, H.; Ishimaru, T.; Yamabe, T.; Tsuji, Y.; Miyamoto, T. Breaking the cycle of HTLV-I transmission via carrier mothers’ milk. Lancet 1987, 2, 158–159. [Google Scholar] [CrossRef]
- Hino, S. Establishment of the milk-borne transmission as a key factor for the peculiar endemicity of human T-lymphotropic virus type 1 (HTLV-1): The ATL Prevention Program Nagasaki. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Gessain, A.; Gallo, R.C.; Franchini, G. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J. Virol. 1992, 66, 2288–2295. [Google Scholar] [CrossRef]
- Tanaka, Y.; Takahashi, Y.; Tanaka, R.; Kodama, A.; Fujii, H.; Hasegawa, A.; Kannagi, M.; Ansari, A.A.; Saito, M. Elimination of human T cell leukemia virus type-1-infected cells by neutralizing and antibody-dependent cellular cytotoxicity-inducing antibodies against human t cell leukemia virus type-1 envelope gp46. AIDS Res. Hum. Retrovir. 2014, 30, 542–552. [Google Scholar] [CrossRef]
- Saito, M.; Tanaka, R.; Fujii, H.; Kodama, A.; Takahashi, Y.; Matsuzaki, T.; Takashima, H.; Tanaka, Y. The neutralizing function of the anti-HTLV-1 antibody is essential in preventing in vivo transmission of HTLV-1 to human T cells in NOD-SCID/γcnull (NOG) mice. Retrovirology 2014, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Shimizu, M.; Miyagi, T.; Kunihiro, M.; Tanaka, R.; Takahashi, Y.; Tanaka, Y. A Potential of an Anti-HTLV-I gp46 Neutralizing Monoclonal Antibody (LAT-27) for Passive Immunization against Both Horizontal and Mother-to-Child Vertical Infection with Human T Cell Leukemia Virus Type-I. Viruses 2016, 8, 41. [Google Scholar] [CrossRef]
- Mitsunaga, M.; Ogawa, M.; Kosaka, N.; Rosenblum, L.T.; Choyke, P.L.; Kobayashi, H. Cancer cell-selective in vivo near infrared photoimmunotherapy targeting specific membrane molecules. Nat. Med. 2011, 17, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Ando, K.; Okuyama, S.; Moriguchi, S.; Ogura, T.; Totoki, S.; Hanaoka, H.; Nagaya, T.; Kokawa, R.; Takakura, H.; et al. Photoinduced Ligand Release from a Silicon Phthalocyanine Dye Conjugated with Monoclonal Antibodies: A Mechanism of Cancer Cell Cytotoxicity after Near-Infrared Photoimmunotherapy. ACS Cent. Sci. 2018, 4, 1559–1569. [Google Scholar] [CrossRef] [PubMed]
- Shirasu, N.; Yamada, H.; Shibaguchi, H.; Kuroki, M.; Kuroki, M. Potent and specific antitumor effect of CEA-targeted photoimmunotherapy. Int. J. Cancer 2014, 135, 2697–2710. [Google Scholar] [CrossRef]
- Sato, K.; Choyke, P.L.; Kobayashi, H. Photoimmunotherapy of gastric cancer peritoneal carcinomatosis in a mouse model. PLoS ONE 2014, 9, e113276. [Google Scholar] [CrossRef] [PubMed]
- Nagaya, T.; Nakamura, Y.; Sato, K.; Harada, T.; Choyke, P.L.; Hodge, J.W.; Schlom, J.; Kobayashi, H. Near infrared photoimmunotherapy with avelumab, an anti-programmed death-ligand 1 (PD-L1) antibody. Oncotarget 2017, 8, 8807–8817. [Google Scholar] [CrossRef]
- Li, F.; Mao, C.; Yeh, S.; Sun, Y.; Xin, J.; Shi, Q.; Ming, X. MRP1-targeted near infrared photoimmunotherapy for drug resistant small cell lung cancer. Int. J. Pharm. 2021, 604, 120760. [Google Scholar] [CrossRef] [PubMed]
- Nagaya, T.; Sato, K.; Harada, T.; Nakamura, Y.; Choyke, P.L.; Kobayashi, H. Near Infrared Photoimmunotherapy Targeting EGFR Positive Triple Negative Breast Cancer: Optimizing the Conjugate-Light Regimen. PLoS ONE 2015, 10, e0136829. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; On, J.; Morita, T.; Suzuki, T.; Okada, Y.; Ono, J.; Evdokiou, A. Molecular Sciences Combination of Near-Infrared Photoimmunotherapy Using Trastuzumab and Small Protein Mimetic for HER2-Positive Breast Cancer. J. Mol. Sci 2021, 22, 12213. [Google Scholar] [CrossRef] [PubMed]
- Hanaoka, H.; Nagaya, T.; Sato, K.; Nakamura, Y.; Watanabe, R.; Harada, T.; Gao, W.; Feng, M.; Phung, Y.; Kim, I.; et al. Glypican-3 targeted human heavy chain antibody as a drug carrier for hepatocellular carcinoma therapy. Mol. Pharm. 2015, 12, 2151–2157. [Google Scholar] [CrossRef]
- Jing, H.; Weidensteiner, C.; Reichardt, W.; Gaedicke, S.; Zhu, X.; Grosu, A.-L.; Kobayashi, H.; Niedermann, G. Imaging and Selective Elimination of Glioblastoma Stem Cells with Theranostic Near-Infrared-Labeled CD133-Specific Antibodies. Theranostics 2016, 6, 862–874. [Google Scholar] [CrossRef]
- Burley, T.A.; Mączyńska, J.; Shah, A.; Szopa, W.; Harrington, K.J.; Boult, J.K.R.; Mrozek-Wilczkiewicz, A.; Vinci, M.; Bamber, J.C.; Kaspera, W.; et al. Near-infrared photoimmunotherapy targeting EGFR-Shedding new light on glioblastoma treatment. Int. J. Cancer 2018, 142, 2363–2374. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Jiang, D.; Ehlerding, E.B.; Barnhart, T.E.; Yang, Y.; Engle, J.W.; Luo, Q.-Y.; Huang, P.; Cai, W. CD146-Targeted Multimodal Image-Guided Photoimmunotherapy of Melanoma. Adv. Sci. 2019, 6, 1801237. [Google Scholar] [CrossRef] [PubMed]
- Nagaya, T.; Nakamura, Y.; Sato, K.; Harada, T.; Choyke, P.L.; Kobayashi, H. Near infrared photoimmunotherapy of B-cell lymphoma. Mol. Oncol. 2016, 10, 1404–1414. [Google Scholar] [CrossRef] [PubMed]
- Heryanto, Y.-D.; Hanaoka, H.; Takahito Nakajima, T.; Yamaguchi, A.; Tsushima, Y. Applying near-infrared photoimmunotherapy to B-cell lymphoma: Comparative evaluation with radioimmunotherapy in tumor xenografts. Ann. Nucl. Med. 2017, 31, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Mitsunaga, M.; Ito, K.; Nishimura, T.; Miyata, H.; Mizunoe, Y.; Kobayashi, H.; Iwase, T. A flexible target-specific anti-infection therapeutic platform that can be applied to different microbial species. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yasui, H.; Takahashi, K.; Taki, S.; Shimizu, M.; Koike, C.; Umeda, K.; Rahman, S.; Akashi, T.; Nguyen, V.S.; Nakagawa, Y.; et al. Near Infrared Photo-Antimicrobial Targeting Therapy for Candida albicans. Adv. Ther. 2021, 4, 2000221. [Google Scholar] [CrossRef]
- Mitsunaga, M.; Ito, K.; Nishimura, T.; Miyata, H.; Miyakawa, K.; Morita, T.; Ryo, A.; Kobayashi, H.; Mizunoe, Y.; Iwase, T. Antimicrobial strategy for targeted elimination of different microbes, including bacterial, fungal and viral pathogens. Commun. Biol. 2022, 5, 647. [Google Scholar] [CrossRef]
- Sadraeian, M.; Bahou, C.; da Cruz, E.F.; Janini, L.M.R.; Sobhie Diaz, R.; Boyle, R.W.; Chudasama, V.; Eduardo Gontijo Guimarães, F. Photoimmunotherapy Using Cationic and Anionic Photosensitizer-Antibody Conjugates against HIV Env-Expressing Cells. Int. J. Mol. Sci. 2020, 21, 9151. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, S.; Masaoka, T.; Sawasaki, T.; Morishita, R.; Iwatani, Y.; Tatsumi, M.; Endo, Y.; Yamamoto, N.; Sugiura, W.; Ryo, A. A cell-free enzymatic activity assay for the evaluation of HIV-1 drug resistance to protease inhibitors. Front. Microbiol. 2015, 6, 1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaoka, Y.; Matsunaga, S.; Jeremiah, S.S.; Nishi, M.; Miyakawa, K.; Morita, T.; Khatun, H.; Shimizu, H.; Okabe, N.; Kimura, H.; et al. Zika virus protease induces caspase-independent pyroptotic cell death by directly cleaving gasdermin D. Biochem. Biophys. Res. Commun. 2021, 534, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, A.; Ogasawara, T.; Matsunaga, S.; Iwasaki, T.; Sawasaki, T.; Endo, Y. Production and partial purification of membrane proteins using a liposome-supplemented wheat cell-free translation system. BMC Biotechnol. 2011, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, S.; Kawakami, S.; Matsuo, I.; Okayama, A.; Tsukagoshi, H.; Kudoh, A.; Matsushima, Y.; Shimizu, H.; Okabe, N.; Hirano, H.; et al. Wheat germ cell-free system-based production of hemagglutinin-neuraminidase glycoprotein of human parainfluenza virus type 3 for generation and characterization of monoclonal antibody. Front. Microbiol. 2014, 5, 208. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Matsuyama, S.; Fukushi, S.; Matsunaga, S.; Matsushima, Y.; Kuroyama, H.; Kimura, H.; Takeda, M.; Chimuro, T.; Ryo, A. Development of Monoclonal Antibody and Diagnostic Test for Middle East Respiratory Syndrome Coronavirus Using Cell-Free Synthesized Nucleocapsid Antigen. Front. Microbiol. 2016, 7, 509. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Miyakawa, K.; Jeremiah, S.S.; Funabashi, R.; Okudela, K.; Kikuchi, S.; Katada, J.; Wada, A.; Takei, T.; Nishi, M.; et al. Highly specific monoclonal antibodies and epitope identification against SARS-CoV-2 nucleocapsid protein for antigen detection tests. Cell Rep. Med. 2021, 2, 100311. [Google Scholar] [CrossRef] [PubMed]
- Alais, S.; Dutartre, H.; Mahieux, R. Quantitative Analysis of Human T-Lymphotropic Virus Type 1 (HTLV-1) Infection Using Co-Culture with Jurkat LTR-Luciferase or Jurkat LTR-GFP Reporter Cells. Methods Mol. Biol. 2017, 1582, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Zeng, L.; Shiraki, H.; Shida, H.; Tozawa, H. Identification of a neutralization epitope on the envelope gp46 antigen of human T cell leukemia virus type I and induction of neutralizing antibody by peptide immunization. J. Immunol. 1991, 147, 354–360. [Google Scholar]
- Seiki, M.; Hattori, S.; Hirayama, Y.; Yoshida, M. Human adult T-cell leukemia virus: Complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA (human leukemia virus/provirus structure/translation frames/polyadenylylation model). Proc. Natl. Acad. Sci. USA 1983, 80, 3618–3622. [Google Scholar] [CrossRef] [PubMed]
- Harbers, M. Wheat germ systems for cell-free protein expression. FEBS Lett. 2014, 588, 2762–2773. [Google Scholar] [CrossRef] [Green Version]
- Anand, S.P.; Grover, J.R.; Tolbert, W.D.; Prévost, J.; Richard, J.; Ding, S.; Baril, S.; Medjahed, H.; Evans, D.T.; Pazgier, M.; et al. Antibody-Induced Internalization of HIV-1 Env Proteins Limits Surface Expression of the Closed Conformation of Env. J. Virol. 2019, 93, e00293-19. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Ogawa, M. Phototoxicity in near-infrared photoimmunotherapy is influenced by the subcellular localization of antibody-IR700. Photodiagnosis Photodyn. Ther. 2020, 31, 101926. [Google Scholar] [CrossRef]
- Endo, Y.; Sawasaki, T. Advances in genome-wide protein expression using the wheat germ cell-free system. Methods Mol. Biol. 2005, 310, 145–167. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Sawasaki, T. Cell-free expression systems for eukaryotic protein production. Curr. Opin. Biotechnol. 2006, 17, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.K.; Datta, A.G. Proteoliposome as the model for the study of membrane-bound enzymes and transport proteins. Mol. Cell. Biochem. 1983, 50, 3–15. [Google Scholar] [CrossRef]
- Takeda, H.; Ogasawara, T.; Ozawa, T.; Muraguchi, A.; Jih, P.-J.; Morishita, R.; Uchigashima, M.; Watanabe, M.; Fujimoto, T.; Iwasaki, T.; et al. Production of monoclonal antibodies against GPCR using cell-free synthesized GPCR antigen and biotinylated liposome-based interaction assay. Sci. Rep. 2015, 5, 11333. [Google Scholar] [CrossRef]
- Ghez, D.; Lepelletier, Y.; Lambert, S.; Fourneau, J.-M.; Blot, V.; Janvier, S.; Arnulf, B.; van Endert, P.M.; Heveker, N.; Pique, C.; et al. Neuropilin-1 Is Involved in Human T-Cell Lymphotropic Virus Type 1 Entry. J. Virol. 2006, 80, 6844–6854. [Google Scholar] [CrossRef]
- Piñon, J.D.; Klasse, P.J.; Jassal, S.R.; Welson, S.; Weber, J.; Brighty, D.W.; Sattentau, Q.J. Human T-Cell Leukemia Virus Type 1 Envelope Glycoprotein gp46 Interacts with Cell Surface Heparan Sulfate Proteoglycans. J. Virol. 2003, 77, 9922–9930. [Google Scholar] [CrossRef]
- Manel, N.; Kim, F.J.; Kinet, S.; Taylor, N.; Sitbon, M.; Battini, J.L. The Ubiquitous Glucose Transporter GLUT-1 Is a Receptor for HTLV. Cell 2003, 115, 449–459. [Google Scholar] [CrossRef]
- Wallin, M.; Ekström, M.; Garoff, H. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 2004, 23, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Zhang, S.; Kronqvist, M.; Wallin, M.; Ekström, M.; Derse, D.; Garoff, H. Intersubunit Disulfide Isomerization Controls Membrane Fusion of Human T-Cell Leukemia Virus Env. J. Virol. 2008, 82, 7135–7143. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.A.; Maerz, A.L.; Bär, S.; Drummer, H.E.; Poumbourios, P. An N-terminal glycine-rich sequence contributes to retrovirus trimer of hairpins stability. Biochem. Biophys. Res. Commun. 2007, 359, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Kuroki, M.; Nakamura, M.; Itoyama, Y.; Tanaka, Y.; Shiraki, H.; Baba, E.; Esaki, T.; Tatsumoto, T.; Nagafuchi, S.; Nakano, S. Identification of new epitopes recognized by human monoclonal antibodies with neutralizing and antibody-dependent cellular cytotoxicity activities specific for human T cell leukemia virus type 1. J. Immunol. 1992, 149, 940–948. [Google Scholar]
- Lavillette, D.; Maurice, M.; Roche, C.; Russell, S.J.; Sitbon, M.; Cosset, F.-L. A Proline-Rich Motif Downstream of the Receptor Binding Domain Modulates Conformation and Fusogenicity of Murine Retroviral Envelopes. J. Virol. 1998, 72, 9955–9965. [Google Scholar] [CrossRef]
- Kobayashi, H.; Furusawa, A.; Rosenberg, A.; Choyke, P.L. Near-infrared photoimmunotherapy of cancer: A new approach that kills cancer cells and enhances anti-cancer host immunity. Int. Immunol. 2021, 33, 7–15. [Google Scholar] [CrossRef]
- Xia, G.-Q.Q.; Lei, T.-R.R.; Yu, T.-B.B.; Zhou, P.-H.H. Nanocarrier-based activation of necroptotic cell death potentiates cancer immunotherapy. Nanoscale 2021, 13, 1220–1230. [Google Scholar] [CrossRef]
- Gaudray, G.; Gachon, F.; Basbous, J.; Biard-Piechaczyk, M.; Devaux, C.; Mesnard, J.-M. The Complementary Strand of the Human T-Cell Leukemia Virus Type 1 RNA Genome Encodes a bZIP Transcription Factor That Down-Regulates Viral Transcription. J. Virol. 2002, 76, 12813–12822. [Google Scholar] [CrossRef]
- Koya, J.; Saito, Y.; Kameda, T.; Kogure, Y.; Yuasa, M.; Nagasaki, J.; McClure, M.B.; Shingaki, S.; Tabata, M.; Tahira, Y.; et al. Single-Cell Analysis of the Multicellular Ecosystem in Viral Carcinogenesis by HTLV-1. Blood Cancer Discov. 2021, 2, 450–467. [Google Scholar] [CrossRef]
- Tezuka, K.; Xun, R.; Tei, M.; Ueno, T.; Tanaka, M.; Takenouchi, N.; Fujisawa, J.I. An animal model of adult T-cell leukemia: Humanized mice with HTLV-1–specific immunity. Blood 2014, 123, 346–355. [Google Scholar] [CrossRef]
- Sagara, Y.; Inoue, Y.; Sagara, Y.; Kashiwagi, S. Involvement of molecular mimicry between human T-cell leukemia virus type 1 gp46 and osteoprotegerin in induction of hypercalcemia. Cancer Sci. 2009, 100, 490–496. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hatayama, Y.; Yamaoka, Y.; Morita, T.; Jeremiah, S.S.; Miyakawa, K.; Nishi, M.; Kimura, Y.; Mitsunaga, M.; Iwase, T.; Kimura, H.; et al. Development of a Monoclonal Antibody Targeting HTLV-1 Envelope gp46 Glycoprotein and Its Application to Near-Infrared Photoimmuno-Antimicrobial Strategy. Viruses 2022, 14, 2153. https://doi.org/10.3390/v14102153
Hatayama Y, Yamaoka Y, Morita T, Jeremiah SS, Miyakawa K, Nishi M, Kimura Y, Mitsunaga M, Iwase T, Kimura H, et al. Development of a Monoclonal Antibody Targeting HTLV-1 Envelope gp46 Glycoprotein and Its Application to Near-Infrared Photoimmuno-Antimicrobial Strategy. Viruses. 2022; 14(10):2153. https://doi.org/10.3390/v14102153
Chicago/Turabian StyleHatayama, Yasuyoshi, Yutaro Yamaoka, Takeshi Morita, Sundararaj Stanleyraj Jeremiah, Kei Miyakawa, Mayuko Nishi, Yayoi Kimura, Makoto Mitsunaga, Tadayuki Iwase, Hirokazu Kimura, and et al. 2022. "Development of a Monoclonal Antibody Targeting HTLV-1 Envelope gp46 Glycoprotein and Its Application to Near-Infrared Photoimmuno-Antimicrobial Strategy" Viruses 14, no. 10: 2153. https://doi.org/10.3390/v14102153





