Dangerous Liaisons: Long-Term Replication with an Extrachromosomal HPV Genome
Abstract
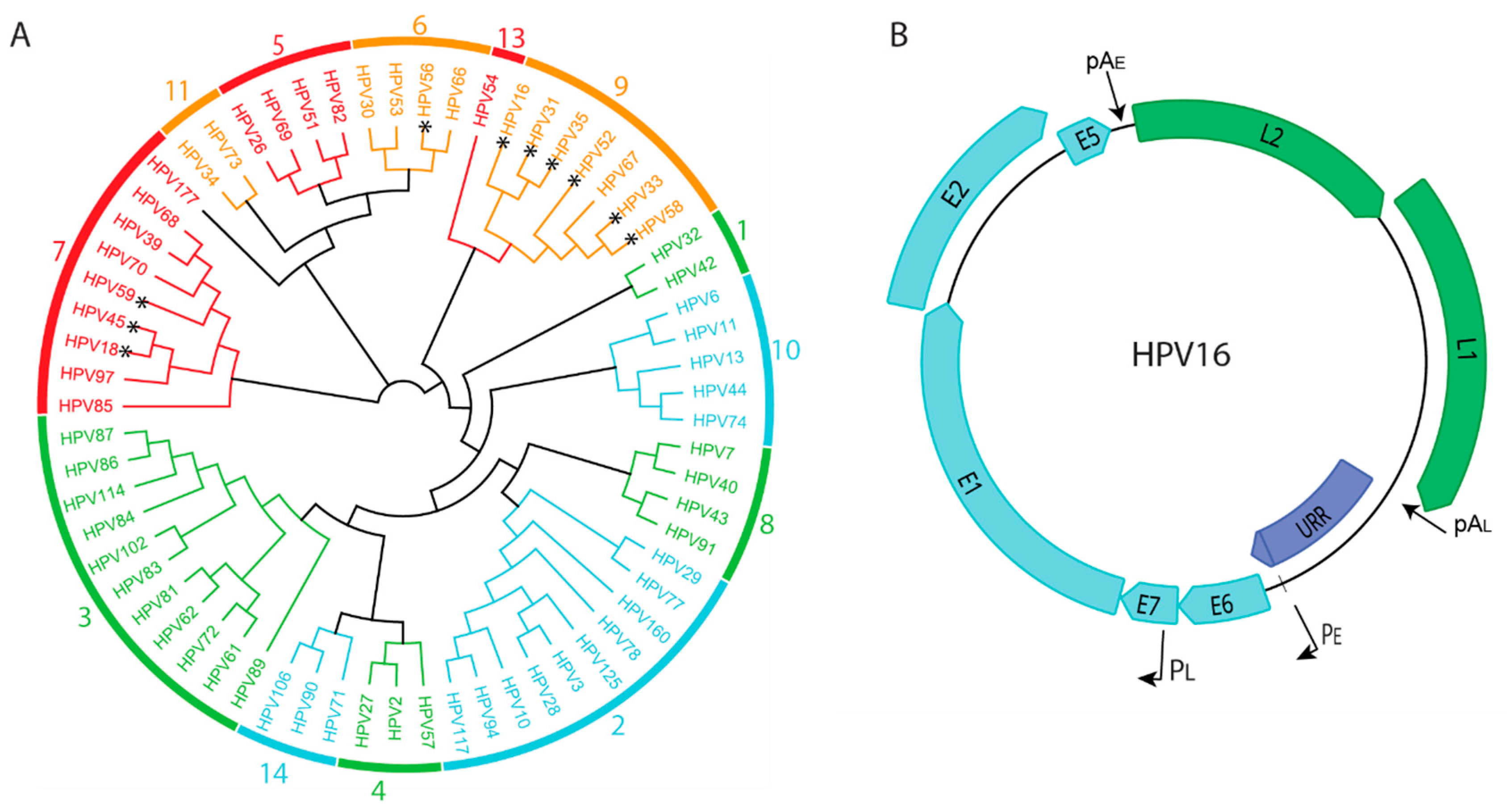
:1. Human Papillomaviruses and Their Association with Human Disease
2. Papillomavirus Genome Organization
3. The Papillomavirus Infectious Cycle
4. The Fate of Viral DNA through the Infectious Cycle
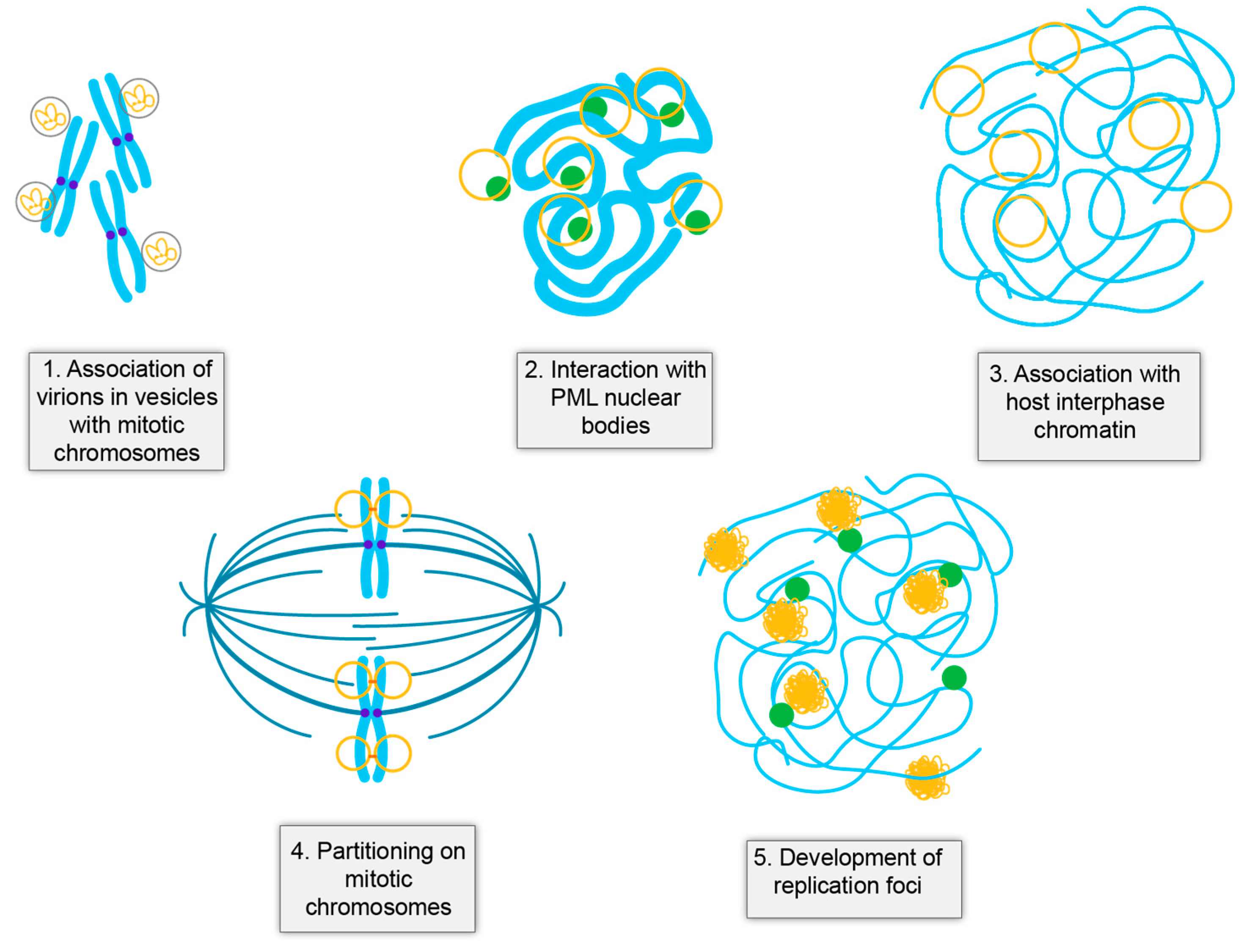
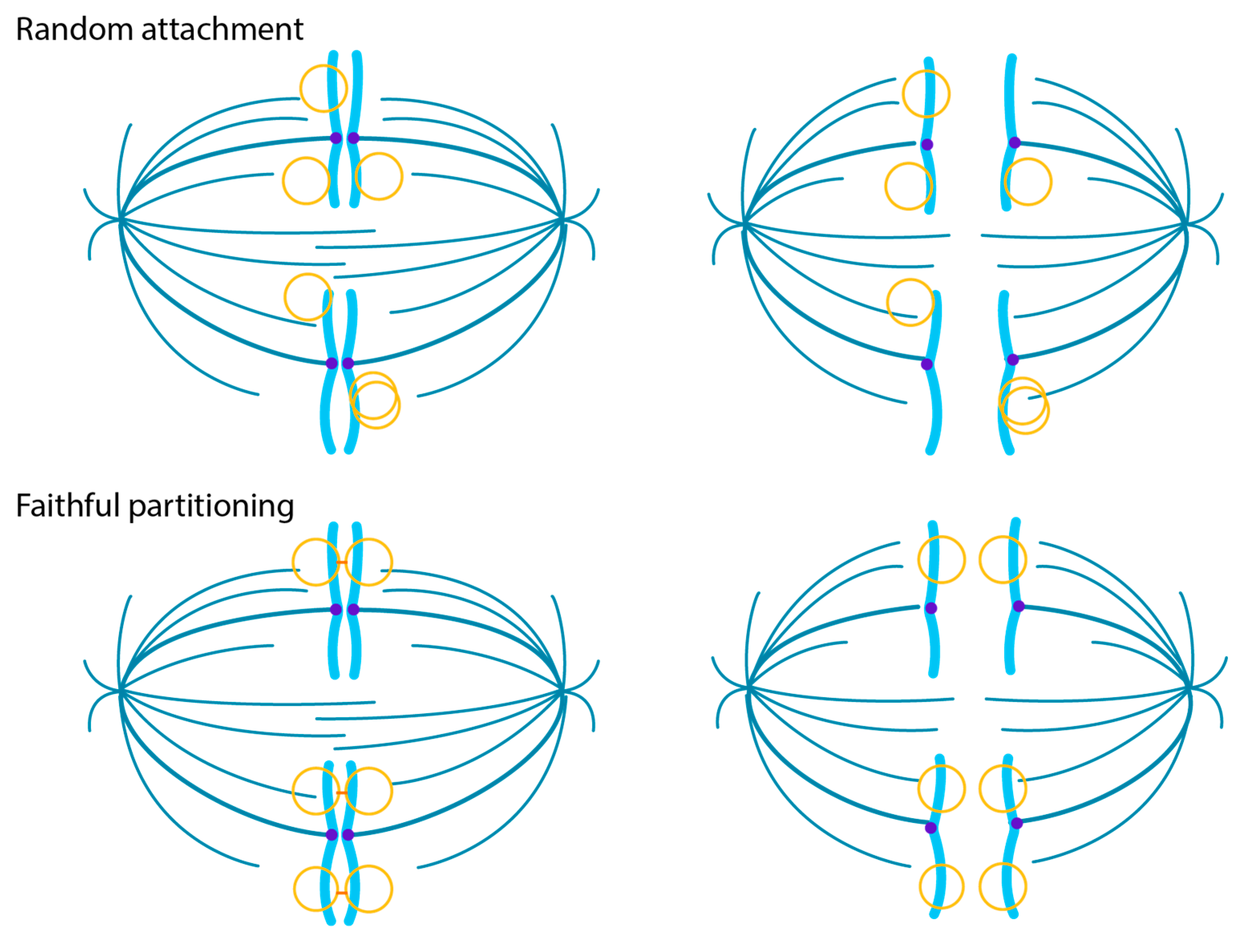
4.1. HPV Virions Associate with Mitotic Chromosomes on Entry
4.2. Establishment of HPV Infection Occurs at PML Nuclear Bodies
4.3. Establishment of Infection
4.4. The Maintenance Phase of Infection
4.5. Viral DNA Amplification in Differentiated Cells
5. Modes of Viral DNA Replication
6. Unique Functions of Oncogenic HPVs
6.1. HPV-Mediated Regulation of Cellular Proliferation and Differentiation
6.2. Evasion of Foreign DNA Recognition in HPV Infected Cells
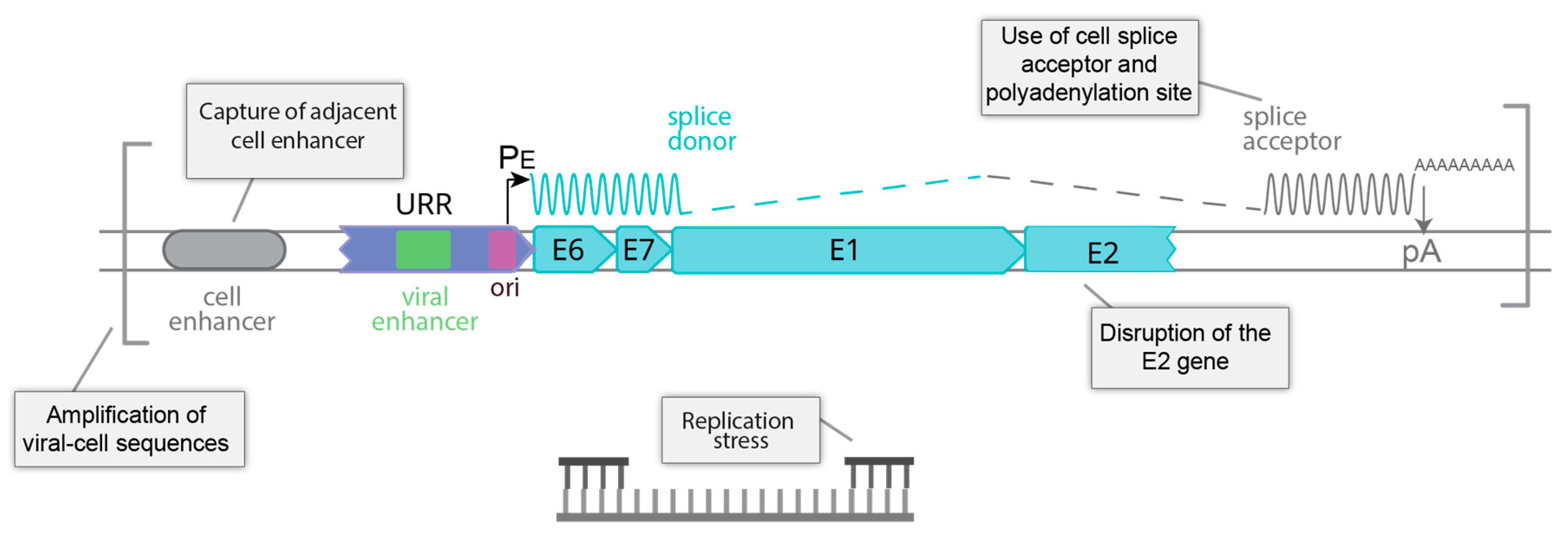
7. HPV Genome Integration
7.1. Integration Is Frequent in HPV Associated Tumors
7.2. Structure of the HPV Genome at Integration Sites
7.3. Landscape of the Cellular Genome at HPV Integration Sites
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Doorslaer, K.; Li, Z.; Xirasagar, S.; Maes, P.; Kaminsky, D.; Liou, D.; Sun, Q.; Kaur, R.; Huyen, Y.; McBride, A.A. The Papillomavirus Episteme: A major update to the papillomavirus sequence database. Nucleic Acids. Res. 2017, 45, D499–D506. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.A. Human papillomaviruses: Diversity, infection and host interactions. Nat. Microbiol. 2021. [Google Scholar] [CrossRef]
- Schiffman, M.; Doorbar, J.; Wentzensen, N.; de Sanjose, S.; Fakhry, C.; Monk, B.J.; Stanley, M.A.; Franceschi, S. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers 2016, 2, 16086. [Google Scholar] [CrossRef] [PubMed]
- Della Fera, A.N.; Warburton, A.; Coursey, T.L.; Khurana, S.; McBride, A.A. Persistent Human Papillomavirus Infection. Viruses 2021, 13, 321. [Google Scholar] [CrossRef] [PubMed]
- Ozbun, M.A. Human papillomavirus type 31b infection of human keratinocytes and the onset of early transcription. J. Virol. 2002, 76, 11291–11300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyeon, D.; Pearce, S.M.; Lank, S.M.; Ahlquist, P.; Lambert, P.F. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009, 5, e1000318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aydin, I.; Weber, S.; Snijder, B.; Samperio Ventayol, P.; Kuhbacher, A.; Becker, M.; Day, P.M.; Schiller, J.T.; Kann, M.; Pelkmans, L.; et al. Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses. PLoS Pathog. 2014, 10, e1004162. [Google Scholar] [CrossRef]
- Calton, C.M.; Bronnimann, M.P.; Manson, A.R.; Li, S.; Chapman, J.A.; Suarez-Berumen, M.; Williamson, T.R.; Molugu, S.K.; Bernal, R.A.; Campos, S.K. Translocation of the papillomavirus L2/vDNA complex across the limiting membrane requires the onset of mitosis. PLoS Pathog. 2017, 13, e1006200. [Google Scholar] [CrossRef] [Green Version]
- DiGiuseppe, S.; Bienkowska-Haba, M.; Guion, L.G.; Sapp, M. Cruising the cellular highways: How human papillomavirus travels from the surface to the nucleus. Virus Res. 2017, 231, 1–9. [Google Scholar] [CrossRef] [Green Version]
- DiGiuseppe, S.; Bienkowska-Haba, M.; Sapp, M. Human Papillomavirus Entry: Hiding in a Bubble. J. Virol. 2016, 90, 8032–8035. [Google Scholar] [CrossRef] [Green Version]
- DiGiuseppe, S.; Luszczek, W.; Keiffer, T.R.; Bienkowska-Haba, M.; Guion, L.G.; Sapp, M.J. Incoming human papillomavirus type 16 genome resides in a vesicular compartment throughout mitosis. Proc. Natl. Acad. Sci. USA 2016, 113, 6289–6294. [Google Scholar] [CrossRef] [Green Version]
- Uhlorn, B.L.; Jackson, R.; Li, S.; Bratton, S.M.; Van Doorslaer, K.; Campos, S.K. Vesicular trafficking permits evasion of cGAS/STING surveillance during initial human papillomavirus infection. PLoS Pathog. 2020, 16, e1009028. [Google Scholar] [CrossRef]
- Aydin, I.; Villalonga-Planells, R.; Greune, L.; Bronnimann, M.P.; Calton, C.M.; Becker, M.; Lai, K.Y.; Campos, S.K.; Schmidt, M.A.; Schelhaas, M. A central region in the minor capsid protein of papillomaviruses facilitates viral genome tethering and membrane penetration for mitotic nuclear entry. PLoS Pathog. 2017, 13, e1006308. [Google Scholar] [CrossRef] [Green Version]
- Day, P.M.; Baker, C.C.; Lowy, D.R.; Schiller, J.T. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl. Acad. Sci. USA 2004, 101, 14252–14257. [Google Scholar] [CrossRef] [Green Version]
- Bienkowska-Haba, M.; Luszczek, W.; Keiffer, T.R.; Guion, L.G.M.; DiGiuseppe, S.; Scott, R.S.; Sapp, M. Incoming human papillomavirus 16 genome is lost in PML protein-deficient HaCaT keratinocytes. Cell Microbiol. 2017, 19, e12708. [Google Scholar] [CrossRef] [Green Version]
- Stepp, W.H.; Meyers, J.M.; McBride, A.A. Sp100 provides intrinsic immunity against human papillomavirus infection. mBio 2013, 4, e00845-13. [Google Scholar] [CrossRef] [Green Version]
- Scherer, M.; Stamminger, T. Emerging Role of PML Nuclear Bodies in Innate Immune Signaling. J. Virol. 2016, 90, 5850–5854. [Google Scholar] [CrossRef] [Green Version]
- Guion, L.G.; Sapp, M. The Role of Promyelocytic Leukemia Nuclear Bodies During HPV Infection. Front. Cell Infect. Microbiol. 2020, 10, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florin, L.; Schafer, F.; Sotlar, K.; Streeck, R.E.; Sapp, M. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology 2002, 295, 97–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.C.; Kappel, C.; Beaudouin, J.; Eils, R.; Spector, D.L. Live cell dynamics of promyelocytic leukemia nuclear bodies upon entry into and exit from mitosis. Mol. Biol. Cell 2008, 19, 3147–3162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guion, L.; Bienkowska-Haba, M.; DiGiuseppe, S.; Florin, L.; Sapp, M. PML nuclear body-residing proteins sequentially associate with HPV genome after infectious nuclear delivery. PLoS Pathog. 2019, 15, e1007590. [Google Scholar] [CrossRef]
- Day, P.M.; Roden, R.B.; Lowy, D.R.; Schiller, J.T. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 1998, 72, 142–150. [Google Scholar] [CrossRef] [Green Version]
- Ching, R.W.; Ahmed, K.; Boutros, P.C.; Penn, L.Z.; Bazett-Jones, D.P. Identifying gene locus associations with promyelocytic leukemia nuclear bodies using immuno-TRAP. J. Cell Biol. 2013, 201, 325–335. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.R.; Munkhjargal, A.; Kim, M.J.; Park, S.Y.; Jung, E.; Ryu, J.H.; Yang, Y.; Lim, J.S.; Kim, Y. The functional roles of PML nuclear bodies in genome maintenance. Mutat. Res. 2018, 809, 99–107. [Google Scholar] [CrossRef]
- Corpet, A.; Kleijwegt, C.; Roubille, S.; Juillard, F.; Jacquet, K.; Texier, P.; Lomonte, P. PML nuclear bodies and chromatin dynamics: Catch me if you can! Nucleic Acids. Res. 2020, 48, 11890–11912. [Google Scholar] [CrossRef]
- Lace, M.J.; Turek, L.P.; Anson, J.R.; Haugen, T.H. Analyzing the Human Papillomavirus (HPV) Life Cycle in Primary Keratinocytes with a Quantitative Colony-Forming Assay. Curr. Protoc. Microbiol. 2014, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Van Doorslaer, K.; Porter, S.; McKinney, C.; Stepp, W.H.; McBride, A.A. Novel recombinant papillomavirus genomes expressing selectable genes. Sci. Rep. 2016, 6, 37782. [Google Scholar] [CrossRef] [Green Version]
- Stanley, M.A.; Browne, H.M.; Appleby, M.; Minson, A.C. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int. J.Cancer 1989, 43, 672–676. [Google Scholar] [CrossRef]
- Bedell, M.A.; Hudson, J.B.; Golub, T.R.; Turyk, M.E.; Hosken, M.; Wilbanks, G.D.; Laimins, L.A. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J.Virol. 1991, 65, 2254–2260. [Google Scholar] [CrossRef] [Green Version]
- Coursey, T.L.; McBride, A.A. Development of Keratinocyte Cell Lines containing Extrachromosomal Human Papillomavirus Genomes. Curr. Protoc. 2021, 1, e235. [Google Scholar] [CrossRef]
- Evans, M.F.; Aliesky, H.A.; Cooper, K. Optimization of biotinyl-tyramide-based in situ hybridization for sensitive background-free applications on formalin-fixed, paraffin-embedded tissue specimens. BMC Clin. Pathol. 2003, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Skiadopoulos, M.H.; McBride, A.A. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 1998, 72, 2079–2088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilves, I.; Kivi, S.; Ustav, M. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 1999, 73, 4404–4412. [Google Scholar] [CrossRef] [Green Version]
- Coursey, T.L.; McBride, A.A. Hitchhiking of Viral Genomes on Cellular Chromosomes. Annu. Rev. Virol. 2019, 6, 275–296. [Google Scholar] [CrossRef]
- Jang, M.K.; Kwon, D.; McBride, A.A. Papillomavirus E2 proteins and the host BRD4 protein associate with transcriptionally active cellular chromatin. J. Virol. 2009, 83, 2592–2600. [Google Scholar] [CrossRef] [Green Version]
- Jang, M.K.; Shen, K.; McBride, A.A. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog. 2014, 10, e1004117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, J.; Croyle, J.L.; Nishimura, A.; Ozato, K.; Howley, P.M. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 2004, 117, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Helfer, C.M.; Yan, J.; You, J. The cellular bromodomain protein Brd4 has multiple functions in E2-mediated papillomavirus transcription activation. Viruses 2014, 6, 3228–3249. [Google Scholar] [CrossRef]
- McBride, A.A.; Warburton, A.; Khurana, S. Multiple Roles of Brd4 in the Infectious Cycle of Human Papillomaviruses. Front. Mol. Biosci. 2021, 8, 727. [Google Scholar] [CrossRef]
- Wu, S.C.; Canarte, V.; Beeravolu, H.; Grace, M.; Sharma, S.; Munger, K. Human Papillomavirus. In Finding How Human Papillomaviruses Alter the Biochemistry and Identity of Infected Epithelial Cells; Jenkins, D., Bosch, F.X., Eds.; Academic Press: San Diego, CA, USA, 2020; pp. 53–65. [Google Scholar]
- Nakahara, T.; Peh, W.L.; Doorbar, J.; Lee, D.; Lambert, P.F. Human papillomavirus type 16 E1^E4 contributes to multiple facets of the papillomavirus life cycle. J. Virol. 2005, 79, 13150–13165. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, N.S.; Wang, H.K.; Broker, T.R.; Chow, L.T. Human papillomavirus (HPV) E7 induces prolonged G2 following S phase reentry in differentiated human keratinocytes. J. Biol. Chem. 2011, 286, 15473–15482. [Google Scholar] [CrossRef] [Green Version]
- Anacker, D.C.; Gautam, D.; Gillespie, K.A.; Chappell, W.H.; Moody, C.A. Productive replication of human papillomavirus 31 requires DNA repair factor Nbs1. J. Virol. 2014, 88, 8528–8544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moody, C.A.; Laimins, L.A. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 2009, 5, e1000605. [Google Scholar] [CrossRef] [Green Version]
- Sakakibara, N.; Mitra, R.; McBride, A.A. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J. Virol. 2011, 85, 8981–8995. [Google Scholar] [CrossRef] [Green Version]
- Fradet-Turcotte, A.; Bergeron-Labrecque, F.; Moody, C.A.; Lehoux, M.; Laimins, L.A.; Archambault, J. Nuclear accumulation of the papillomavirus E1 helicase blocks S-phase progression and triggers an ATM-dependent DNA damage response. J. Virol. 2011, 85, 8996–9012. [Google Scholar] [CrossRef] [Green Version]
- Reinson, T.; Toots, M.; Kadaja, M.; Pipitch, R.; Allik, M.; Ustav, E.; Ustav, M. Engagement of the ATR-dependent DNA damage response at the human papillomavirus 18 replication centers during the initial amplification. J. Virol. 2013, 87, 951–964. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, K.A.; Mehta, K.P.; Laimins, L.A.; Moody, C.A. Human papillomaviruses recruit cellular DNA repair and homologous recombination factors to viral replication centers. J. Virol. 2012, 86, 9520–9526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chappell, W.H.; Gautam, D.; Ok, S.T.; Johnson, B.A.; Anacker, D.C.; Moody, C.A. Homologous Recombination Repair Factors Rad51 and BRCA1 Are Necessary for Productive Replication of Human Papillomavirus 31. J. Virol. 2015, 90, 2639–2652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moody, C. Mechanisms by which HPV Induces a Replication Competent Environment in Differentiating Keratinocytes. Viruses 2017, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Sitz, J.; Blanchet, S.A.; Gameiro, S.F.; Biquand, E.; Morgan, T.M.; Galloy, M.; Dessapt, J.; Lavoie, E.G.; Blondeau, A.; Smith, B.C.; et al. Human papillomavirus E7 oncoprotein targets RNF168 to hijack the host DNA damage response. Proc. Natl. Acad. Sci. USA 2019, 116, 19552–19562. [Google Scholar] [CrossRef] [Green Version]
- Sakakibara, N.; Chen, D.; McBride, A.A. Papillomaviruses use recombination-dependent replication to vegetatively amplify their genomes in differentiated cells. PLoS Pathog. 2013, 9, e1003321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, K.; Laimins, L. Human Papillomaviruses Preferentially Recruit DNA Repair Factors to Viral Genomes for Rapid Repair and Amplification. mBio. 2018, 9, e00064-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debatisse, M.; Rosselli, F. A journey with common fragile sites: From S phase to telophase. Genes Chromosomes Cancer 2019, 58, 305–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ustav, M.; Ustav, E.; Szymanski, P.; Stenlund, A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991, 10, 4321–4329. [Google Scholar] [CrossRef] [PubMed]
- Ustav, M.; Stenlund, A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991, 10, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Piirsoo, M.; Ustav, E.; Mandel, T.; Stenlund, A.; Ustav, M. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 1996, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Lin, B.Y.; Jin, G.; Wheeler, C.G.; Ma, T.; Harper, J.W.; Broker, T.R.; Chow, L.T. Cyclin/CDK regulates the nucleocytoplasmic localization of the human papillomavirus E1 DNA helicase. J.Virol. 2004, 78, 13954–13965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fradet-Turcotte, A.; Moody, C.; Laimins, L.A.; Archambault, J. Nuclear export of human papillomavirus type 31 E1 is regulated by Cdk2 phosphorylation and required for viral genome maintenance. J.Virol. 2010, 84, 11747–11760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Lambert, P.F. E1 protein of bovine papillomavirus 1 is not required for the maintenance of viral plasmid DNA replication. Virology 2002, 293, 10–14. [Google Scholar] [CrossRef] [Green Version]
- Murakami, I.; Egawa, N.; Griffin, H.; Yin, W.; Kranjec, C.; Nakahara, T.; Kiyono, T.; Doorbar, J. Roles for E1-independent replication and E6-mediated p53 degradation during low-risk and high-risk human papillomavirus genome maintenance. PLoS Pathog. 2019, 15, e1007755. [Google Scholar] [CrossRef] [Green Version]
- Egawa, N.; Nakahara, T.; Ohno, S.; Narisawa-Saito, M.; Yugawa, T.; Fujita, M.; Yamato, K.; Natori, Y.; Kiyono, T. The E1 protein of human papillomavirus type 16 is dispensable for maintenance replication of the viral genome. J. Virol. 2012, 86, 3276–3283. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, R.; Hirt, B.; Bechtold, V.; Beard, P.; Raj, K. Different modes of human papillomavirus DNA replication during maintenance. J. Virol. 2006, 80, 4431–4439. [Google Scholar] [CrossRef] [Green Version]
- Flores, E.R.; Lambert, P.F. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J.Virol. 1997, 71, 7167–7179. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Bellanger, S.; Zhang, W.; Lim, D.; Low, J.; Lunny, D.; Thierry, F. HPV16 E2 is an immediate early marker of viral infection, preceding E7 expression in precursor structures of cervical carcinoma. Cancer Res. 2010, 70, 5316–5325. [Google Scholar] [CrossRef] [Green Version]
- Klumpp, D.J.; Laimins, L.A. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology 1999, 257, 239–246. [Google Scholar] [CrossRef] [Green Version]
- Johansson, C.; Schwartz, S. Regulation of human papillomavirus gene expression by splicing and polyadenylation. Nat. Rev. Microbiol. 2013, 11, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Cheng, S.; Iovane, A.; Laimins, L.A. STAT-5 Regulates Transcription of the Topoisomerase IIbeta-Binding Protein 1 (TopBP1) Gene To Activate the ATR Pathway and Promote Human Papillomavirus Replication. mBio 2015, 6, e02006-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.; Li, Y.; Kaminski, P.J.; Andrade, J.; Laimins, L.A. Pathogenesis of Human Papillomaviruses Requires the ATR/p62 Autophagy-Related Pathway. mBio 2020, 11, e01628-20. [Google Scholar] [CrossRef]
- Mehta, K.; Gunasekharan, V.; Satsuka, A.; Laimins, L.A. Human papillomaviruses activate and recruit SMC1 cohesin proteins for the differentiation-dependent life cycle through association with CTCF insulators. PLoS Pathog. 2015, 11, e1004763. [Google Scholar] [CrossRef]
- Gao, G.; Johnson, S.H.; Vasmatzis, G.; Pauley, C.E.; Tombers, N.M.; Kasperbauer, J.L.; Smith, D.I. Common fragile sites (CFS) and extremely large CFS genes are targets for human papillomavirus integrations and chromosome rearrangements in oropharyngeal squamous cell carcinoma. Genes Chromosomes Cancer 2017, 56, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Thorland, E.C.; Myers, S.L.; Gostout, B.S.; Smith, D.I. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene 2003, 22, 1225–1237. [Google Scholar] [CrossRef] [Green Version]
- Thorland, E.C.; Myers, S.L.; Persing, D.H.; Sarkar, G.; McGovern, R.M.; Gostout, B.S.; Smith, D.I. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 2000, 60, 5916–5921. [Google Scholar]
- Smith, P.P.; Friedman, C.L.; Bryant, E.M.; McDougall, J.K. Viral integration and fragile sites in human papillomavirus-immortalized human keratinocyte cell lines. Genes Chromosomes Cancer 1992, 5, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Popescu, N.C.; DiPaolo, J.A. Integration of human papillomavirus 16 DNA and genomic rearrangements in immortalized human keratinocyte lines. Cancer Res. 1990, 50, 1316–1323. [Google Scholar]
- Doorbar, J.; Griffin, H. Refining our understanding of cervical neoplasia and its cellular origins. Papillomavirus Res. 2019, 7, 176–179. [Google Scholar] [CrossRef]
- Doorbar, J. Host control of human papillomavirus infection and disease. Best Pract. Res. Clin. Obs. Gynaecol. 2018, 47, 27–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egawa, N.; Doorbar, J. The low-risk papillomaviruses. Virus Res. 2017, 231, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.S.; Chang, Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer 2010, 10, 878–889. [Google Scholar] [CrossRef] [Green Version]
- Ganti, K.; Broniarczyk, J.; Manoubi, W.; Massimi, P.; Mittal, S.; Pim, D.; Szalmas, A.; Thatte, J.; Thomas, M.; Tomaic, V.; et al. The Human Papillomavirus E6 PDZ Binding Motif: From Life Cycle to Malignancy. Viruses 2015, 7, 3530–3551. [Google Scholar] [CrossRef] [Green Version]
- Klingelhutz, A.J.; Foster, S.A.; McDougall, J.K. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 1996, 380, 79–82. [Google Scholar] [CrossRef]
- Day, P.M.; Weisberg, A.S.; Thompson, C.D.; Hughes, M.M.; Pang, Y.Y.; Lowy, D.R.; Schiller, J.T. Human Papillomavirus 16 Capsids Mediate Nuclear Entry during Infection. J. Virol. 2019, 93, e00454-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiGiuseppe, S.; Keiffer, T.R.; Bienkowska-Haba, M.; Luszczek, W.; Guion, L.G.; Muller, M.; Sapp, M. Topography of the Human Papillomavirus Minor Capsid Protein L2 during Vesicular Trafficking of Infectious Entry. J. Virol. 2015, 89, 10442–10452. [Google Scholar] [CrossRef] [Green Version]
- Favre, M.; Breitburd, F.; Croissant, O.; Orth, G. Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J.Virol. 1977, 21, 1205–1209. [Google Scholar] [CrossRef] [Green Version]
- Kujirai, T.; Zierhut, C.; Takizawa, Y.; Kim, R.; Negishi, L.; Uruma, N.; Hirai, S.; Funabiki, H.; Kurumizaka, H. Structural basis for the inhibition of cGAS by nucleosomes. Science 2020, 370, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.A.; Spangler, C.J.; Strauss, J.D.; Cesmat, A.P.; Liu, P.; McGinty, R.K.; Zhang, Q. Structural basis of nucleosome-dependent cGAS inhibition. Science 2020, 370, 450–454. [Google Scholar] [CrossRef]
- Pathare, G.R.; Decout, A.; Gluck, S.; Cavadini, S.; Makasheva, K.; Hovius, R.; Kempf, G.; Weiss, J.; Kozicka, Z.; Guey, B.; et al. Structural mechanism of cGAS inhibition by the nucleosome. Nature 2020, 587, 668–672. [Google Scholar] [CrossRef]
- Habiger, C.; Jager, G.; Walter, M.; Iftner, T.; Stubenrauch, F. Interferon Kappa Inhibits Human Papillomavirus 31 Transcription by Inducing Sp100 Proteins. J. Virol. 2015, 90, 694–704. [Google Scholar] [CrossRef] [Green Version]
- Lo Cigno, I.; De Andrea, M.; Borgogna, C.; Albertini, S.; Landini, M.M.; Peretti, A.; Johnson, K.E.; Chandran, B.; Landolfo, S.; Gariglio, M. The Nuclear DNA Sensor IFI16 Acts as a Restriction Factor for Human Papillomavirus Replication through Epigenetic Modifications of the Viral Promoters. J. Virol. 2015, 89, 7506–7520. [Google Scholar] [CrossRef] [Green Version]
- Stepp, W.H.; Stamos, J.D.; Khurana, S.; Warburton, A.; McBride, A.A. Sp100 colocalizes with HPV replication foci and restricts the productive stage of the infectious cycle. PLoS Pathog. 2017, 13, e1006660. [Google Scholar] [CrossRef] [Green Version]
- Scott, M.L.; Woodby, B.L.; Ulicny, J.; Raikhy, G.; Orr, A.W.; Songock, W.K.; Bodily, J.M. Human Papillomavirus 16 E5 Inhibits Interferon Signaling and Supports Episomal Viral Maintenance. J. Virol. 2020, 94, e01582-19. [Google Scholar] [CrossRef]
- Hong, S.; Laimins, L.A. Manipulation of the innate immune response by human papillomaviruses. Virus Res. 2017, 231, 34–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo Cigno, I.; Calati, F.; Albertini, S.; Gariglio, M. Subversion of Host Innate Immunity by Human Papillomavirus Oncoproteins. Pathogens 2020, 9, 292. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network, Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [CrossRef]
- Kristiansen, E.; Jenkins, A.; Holm, R. Coexistence of episomal and integrated HPV16 DNA in squamous cell carcinoma of the cervix. J. Clin. Pathol. 1994, 47, 253–256. [Google Scholar] [CrossRef] [Green Version]
- Parfenov, M.; Pedamallu, C.S.; Gehlenborg, N.; Freeman, S.S.; Danilova, L.; Bristow, C.A.; Lee, S.; Hadjipanayis, A.G.; Ivanova, E.V.; Wilkerson, M. Det al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 15544–15549. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, E.; Freese, U.K.; Gissmann, L.; Mayer, W.; Roggenbuck, B.; Stremlau, A.; zur Hausen, H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 1985, 314, 111–114. [Google Scholar] [CrossRef]
- Wentzensen, N.; Vinokurova, S.; von Knebel Doeberitz, M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004, 64, 3878–3884. [Google Scholar] [CrossRef] [Green Version]
- Nulton, T.J.; Olex, A.L.; Dozmorov, M.; Morgan, I.M.; Windle, B. Analysis of the cancer genome atlas sequencing data reveals novel properties of the human papillomavirus 16 genome in head and neck squamous cell carcinoma. Oncotarget 2017, 8, 17684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, B.; Chotewutmontri, S.; Wolf, S.; Klos, U.; Schmitz, M.; Durst, M.; Schwarz, E. Multiplex Identification of Human Papillomavirus 16 DNA Integration Sites in Cervical Carcinomas. PLoS ONE 2013, 8, e66693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warburton, A.; Markowitz, T.E.; Katz, J.P.; Pipas, J.M.; McBride, A.A. Recurrent Integration of Human Papillomavirus Genomes at Transcriptional Regulatory Hubs. BioRxiv 2021. [Google Scholar] [CrossRef]
- Shukla, S.; Mahata, S.; Shishodia, G.; Pande, S.; Verma, G.; Hedau, S.; Bhambhani, S.; Kumari, A.; Batra, S.; Basir, S.F.; et al. Physical state & copy number of high risk human papillomavirus type 16 DNA in progression of cervical cancer. Indian J. Med. Res. 2014, 139, 531–543. [Google Scholar] [PubMed]
- Jeon, S.; Allenhoffmann, B.L.; Lambert, P.F. Integration of Human Papillomavirus Type-16 into the Human Genome Correlates with a Selective Growth Advantage of Cells. J. Virol. 1995, 69, 2989–2997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Majerciak, V.; Xue, X.Y.; Uberoi, A.; Lobanov, A.; Chen, X.; Cam, M.; Hughes, S.H.; Lambert, P.F.; Zheng, Z.M. Mouse papillomavirus type 1 (MmuPV1) DNA is frequently integrated in benign tumors by microhomology-mediated end-joining. PLoS Pathog. 2021, 17, e1009812. [Google Scholar] [CrossRef]
- Goodwin, E.C.; Yang, E.; Lee, C.J.; Lee, H.W.; DiMaio, D.; Hwang, E.S. Rapid induction of senescence in human cervical carcinoma cells. Proc. Natl. Acad. Sci. USA 2000, 97, 10978–10983. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, E.C.; DiMaio, D. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. USA 2000, 97, 12513–12518. [Google Scholar] [CrossRef] [Green Version]
- Wentzensen, N.; Ridder, R.; Klaes, R.; Vinokurova, S.; Schaefer, U.; Doeberitz, M. Characterization of viral-cellular fusion transcripts in a large series of HPV16 and 18 positive anogenital lesions. Oncogene 2002, 21, 419–426. [Google Scholar] [CrossRef] [Green Version]
- Jeon, S.; Lambert, P.F. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: Implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 1995, 92, 1654–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thierry, F.; Heard, J.M.; Dartmann, K.; Yaniv, M. Characterization of a transcriptional promoter of human papillomavirus 18 and modulation of its expression by simian virus 40 and adenovirus early antigens. J.Virol. 1987, 61, 134–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, T.W.; Liu, S.S.; Leung, R.C.; Chu, M.M.; Cheung, A.N.; Ngan, H.Y. HPV 16 E2 binding sites 1 and 2 become more methylated than E2 binding site 4 during cervical carcinogenesis. J. Med. Virol. 2015, 87, 1022–1033. [Google Scholar] [CrossRef]
- Chaiwongkot, A.; Vinokurova, S.; Pientong, C.; Ekalaksananan, T.; Kongyingyoes, B.; Kleebkaow, P.; Chumworathayi, B.; Patarapadungkit, N.; Reuschenbach, M.; von Knebel Doeberitz, M. Differential methylation of E2 binding sites in episomal and integrated HPV 16 genomes in preinvasive and invasive cervical lesions. Int. J. Cancer. 2013, 132, 2087–2094. [Google Scholar] [CrossRef]
- Warburton, A.; Redmond, C.J.; Dooley, K.E.; Fu, H.; Gillison, M.L.; Akagi, K.; Symer, D.E.; Aladjem, M.I.; McBride, A.A. HPV integration hijacks and multimerizes a cellular enhancer to generate a viral-cellular super-enhancer that drives high viral oncogene expression. PLoS Genet. 2018, 14, e1007179. [Google Scholar] [CrossRef]
- McBride, A.A.; Warburton, A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017, 13, e1006211. [Google Scholar] [CrossRef] [Green Version]
- Dooley, K.E.; Warburton, A.; McBride, A.A. Tandemly Integrated HPV16 Can Form a Brd4-Dependent Super-Enhancer-Like Element That Drives Transcription of Viral Oncogenes. mBio 2016, 7, e01446-16. [Google Scholar] [CrossRef] [Green Version]
- Herrick, J.; Conti, C.; Teissier, S.; Thierry, F.; Couturier, J.; Sastre-Garau, X.; Favre, M.; Orth, G.; Bensimon, A. Genomic organization of amplified MYC genes suggests distinct mechanisms of amplification in tumorigenesis. Cancer Res. 2005, 65, 1174–1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014, 24, 185–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, I.M.; DiNardo, L.J.; Windle, B. Integration of Human Papillomavirus Genomes in Head and Neck Cancer: Is It Time to Consider a Paradigm Shift? Viruses 2017, 9, 208. [Google Scholar] [CrossRef] [PubMed]
- Kadaja, M.; Isok-Paas, H.; Laos, T.; Ustav, E.; Ustav, M. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog. 2009, 5, e1000397. [Google Scholar] [CrossRef]
- Kadaja, M.; Sumerina, A.; Verst, T.; Ojarand, M.; Ustav, E.; Ustav, M. Genomic instability of the host cell induced by the human papillomavirus replication machinery. EMBO J. 2007, 26, 2180–2191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, T.E.; Arlt, M.F.; Park, S.H.; Rajendran, S.; Paulsen, M.; Ljungman, M.; Glover, T.W. Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome Res. 2015, 25, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Kelley, D.Z.; Flam, E.L.; Izumchenko, E.; Danilova, L.V.; Wulf, H.A.; Guo, T.; Singman, D.A.; Afsari, B.; Skaist, A.M.; Considine, M.; et al. Integrated Analysis of Whole-Genome ChIP-Seq and RNA-Seq Data of Primary Head and Neck Tumor Samples Associates HPV Integration Sites with Open Chromatin Marks. Cancer Res. 2017, 77, 6538–6550. [Google Scholar] [CrossRef] [Green Version]
- Christiansen, I.K.; Sandve, G.K.; Schmitz, M.; Durst, M.; Hovig, E. Transcriptionally active regions are the preferred targets for chromosomal HPV integration in cervical carcinogenesis. PLoS ONE 2015, 10, e0119566. [Google Scholar]
- Bodelon, C.; Untereiner, M.E.; Machiela, M.J.; Vinokurova, S.; Wentzensen, N. Genomic characterization of viral integration sites in HPV-related cancers. Int. J. Cancer 2016, 139, 2001–2011. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.; Lameiras, S.; Jeannot, E.; Marie, Y.; Castera, L.; Sastre-Garau, X.; Nicolas, A. Mechanistic signatures of HPV insertions in cervical carcinomas. NPJ Genom. Med. 2016, 1, 16004. [Google Scholar] [CrossRef] [Green Version]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-enhancers in the control of cell identity and disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef] [Green Version]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groves, I.J.; Drane, E.L.A.; Michalski, M.; Monahan, J.M.; Scarpini, C.G.; Smith, S.P.; Bussotti, G.; Várnai, C.; Schoenfelder, S.; Fraser, P.; et al. Three-dimensional interactions between integrated HPV genomes and cellular chromatin dysregulate host gene expression in early cervical carcinogenesis. PLOS Pathog. 2021, 17, e1009875. [Google Scholar] [CrossRef]
- Cao, C.; Hong, P.; Huang, X.; Lin, D.; Cao, G.; Wang, L.; Feng, B.; Wu, P.; Shen, H.; Xu, Q.; et al. HPV-CCDC106 integration alters local chromosome architecture and hijacks an enhancer by 3D genome structure remodeling in cervical cancer. J. Genet. Genom. 2020, 47, 437. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Warburton, A.; Della Fera, A.N.; McBride, A.A. Dangerous Liaisons: Long-Term Replication with an Extrachromosomal HPV Genome. Viruses 2021, 13, 1846. https://doi.org/10.3390/v13091846
Warburton A, Della Fera AN, McBride AA. Dangerous Liaisons: Long-Term Replication with an Extrachromosomal HPV Genome. Viruses. 2021; 13(9):1846. https://doi.org/10.3390/v13091846
Chicago/Turabian StyleWarburton, Alix, Ashley N. Della Fera, and Alison A. McBride. 2021. "Dangerous Liaisons: Long-Term Replication with an Extrachromosomal HPV Genome" Viruses 13, no. 9: 1846. https://doi.org/10.3390/v13091846





