CpGV-M Replication in Type I Resistant Insects: Helper Virus and Order of Ingestion Are Important
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Viruses
2.3. Infection Categories and Virus Processing
2.3.1. Order of Infection
2.3.2. Delay of Ingestion
2.3.3. Helper Effect Specificity
2.4. Quantitative Polymerase Chain Reaction (qPCR) and High-Resolution Melting (HRM) Analysis
3. Results
3.1. Importance of the Order of Infection with CpGV-M and CpGV-R5
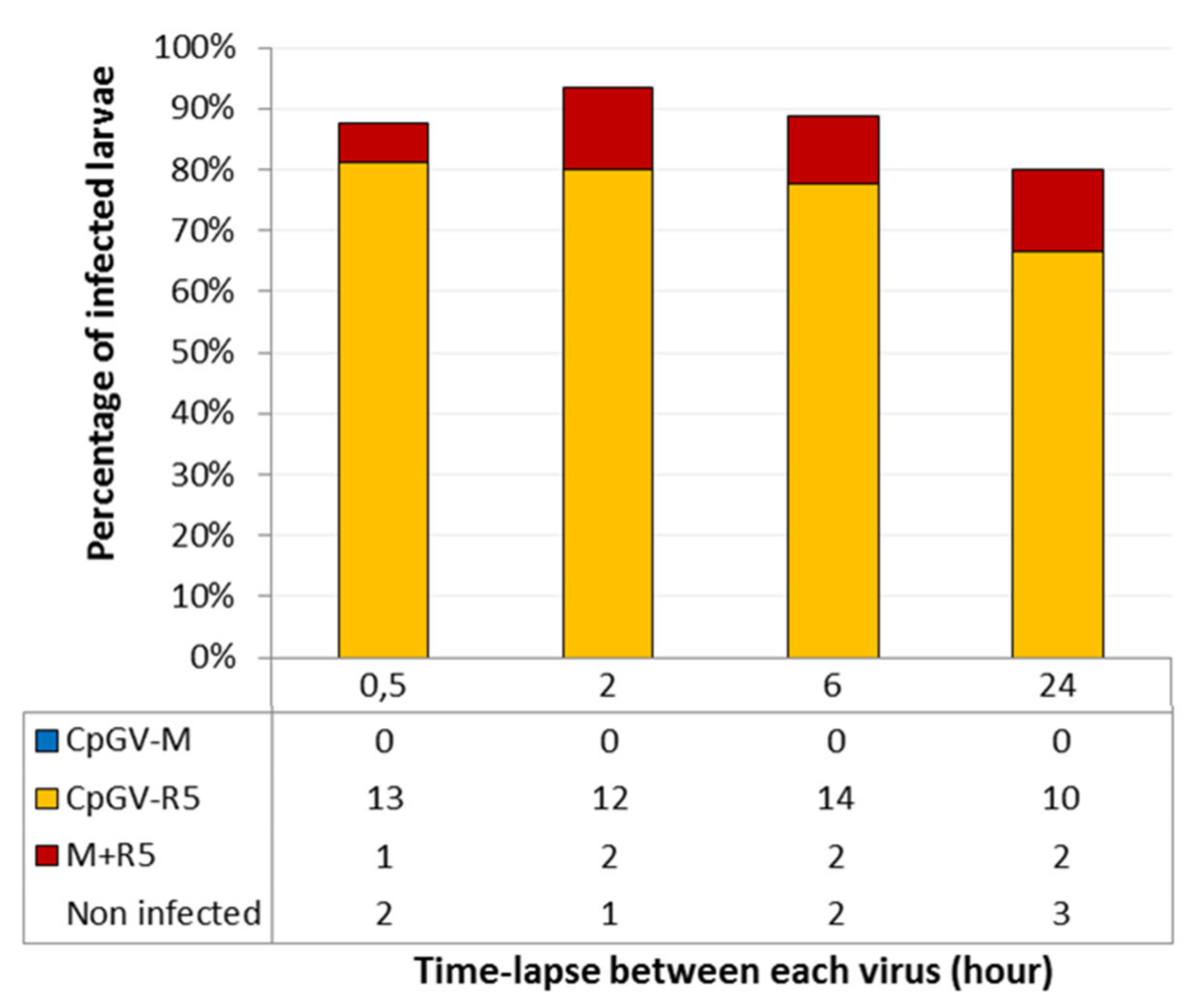
3.2. Delay between the Ingestion of the Different Genotypes
3.3. Double Infections Using CrpeNPV
4. Discussion
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Moscardi, F. Assessment of the application of baculoviruses for control of Lepidoptera. Annu. Rev. Entomol. 1999, 44, 257–289. [Google Scholar] [CrossRef]
- Das, S.; Goswami, A.; Debnath, N. Application of baculoviruses as biopesticides and the possibilities of nanoparticle mediated delivery. In Nano-Biopesticides Today and Future Perspectives; Elsevier: Amsterdam, The Netherlands, 2019; Chapter 11; pp. 261–280. [Google Scholar]
- Federici, B.A. Ultrastructure of baculoviruses. In The Biology of Baculoviruses; Granados, R.R., Federici, B.A., Eds.; CRC Press: Boca Raton, FL, USA, 1986; Volume 1, Chapter 3; pp. 61–88. [Google Scholar]
- Falcon, L.A.; Hess, R.T. Electron microscope observations of multiple occluded virions in the granulosis virus of the codling moth, Cydia Pomonella. J. Invertebr. Pathol. 1985, 45, 356–359. [Google Scholar] [CrossRef]
- Sanjuan, R. Collective infectious units in viruses. Trends Microbiol. 2017, 25, 402–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takatsuka, J.; Okuno, S.; Nakai, M.; Kunimi, Y. Genetic and biological comparisons of ten geographic isolates of a nucleopolyhedrovirus that infects Spodoptera litura (Lepidoptera: Noctuidae). Biol. Control 2003, 26, 32–39. [Google Scholar] [CrossRef]
- Kamiya, K.; Zhu, J.; Murata, M.; Laviña-Caoili, B.A.; Ikeda, M.; Kobayashi, M.; Kawamura, S. Cloning and comparative characterization of three distinct nucleopolyhedroviruses isolated from the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae) in Japan. Biol. Control 2004, 31, 38–48. [Google Scholar] [CrossRef]
- Clavijo, G.; Williams, T.; Muñoz, D.; Caballero, P.; Lopez-Ferber, M. Mixed genotype transmission bodies and virions contribute to the maintenance of diversity in an insect virus. Proc. R. Soc. 2009, 277, 943–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beperet, I.; Simón, O.; López-Ferber, M.; van Lent, J.; Williams, T.; Caballero, P. Mixtures of insect-pathogenic viruses in a single virion: Towards the development of custom-designed Insecticides. Appl. Environ. Microbiol. 2020, 87, e02180-20. [Google Scholar] [CrossRef] [PubMed]
- Escribano, A.; Williams, T.; Goulson, D.; Cave, R.D.; Chapman, J.W.; Caballero, P. Selection of a nucleopolyhedrovirus for control of Spodoptera frugiperda (Lepidoptera: Noctuidae): Structural, genetic, and biological comparison of four isolates from the Americas. J. Econ. Entomol. 1999, 92, 1079–1085. [Google Scholar] [CrossRef] [PubMed]
- Kroschel, J.; Fritsch, E.; Huber, J. Biological control of the potato tuber moth (Phthorimaea operculella Zeller) in the Republic of Yemen using granulosis virus: Biochemical characterization, pathogenicity and stability of the virus. Biocontrol Sci. Technol. 1996, 6, 207–216. [Google Scholar] [CrossRef]
- Graham, R.I.; Tyne, W.I.; Possee, R.D.; Sait, S.M.; Hails, R.S. Genetically variable nucleopolyhedroviruses isolated from spatially separate populations of the winter moth Operophtera brumata (Lepidoptera: Geometridae) in Orkney. J. Invertebr. Pathol. 2004, 87, 29–38. [Google Scholar] [CrossRef]
- Cory, J.S.; Green, B.M.; Paul, R.K.; Hunter-Fujita, F. Genotypic and phenotypic diversity of a baculovirus population within an individual insect host. J. Invertebr. Pathol. 2005, 89, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, D.J.; Vanbergen, A.J.; Watt, A.D.; Hails, R.S.; Cory, J.S. Phenotypic variation between naturally co-existing genotypes of a Lepidopteran baculovirus. Evol. Ecol. Res. 2001, 3, 687–701. [Google Scholar]
- Hinsberger, A. Structuration des Populations Virales Chez les Baculovirus. Importance de L’infection Multiple. Ph.D. Thesis, IMT Mines Alès, Alès, France, 2020. [Google Scholar]
- Simón, O.; Williams, T.; López-Ferber, M.; Caballero, P. Functional importance of deletion mutant genotypes in an insect nucleopolyhedrovirus population. Appl. Environ. Microbiol. 2005, 71, 4254–4262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Ferber, M.; Simón, O.; Williams, T.; Caballero, P. Defective or effective? Mutualistic interactions between virus genotypes. Proc. R. Soc. B Biol. Sci. 2003, 270, 2249–2255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simón, O.; Williams, T.; Caballero, P.; López-Ferber, M. Dynamics of deletion genotypes in an experimental insect virus population. Proc. R. Soc. B Biol. Sci. 2006, 273, 783–790. [Google Scholar] [CrossRef] [Green Version]
- Espinel-Correal, C.; López-Ferber, M.; Zeddam, J.-L.; Villamizar, L.; Gómez, J.; Cotes, A.M.; Léry, X. Experimental mixtures of Phthorimaea operculella granulovirus isolates provide high biological efficacy on both Phthorimaea operculella and Tecia solanivora (Lepidoptera: Gelechiidae). J. Invertebr. Pathol. 2012, 110, 375–381. [Google Scholar] [CrossRef]
- Espinel-Correal, C.; Léry, X.; Villamizar, L.; Gómez, J.; Zeddam, J.L.; Cotes, A.M.; López-Ferber, M. Genetic and biological analysis of colombian Phthorimaea operculella granulovirus isolated from Tecia solanivora (Lepidoptera: Gelechiidae). Appl. Environ. Microbiol. 2010, 76, 7617–7625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritsch, E.; Undorf-Spahn, K.; Kienzle, J.; Zebitz, C.; Huber, J. Codling moth granulovirus: First indication of variations in the susceptibility of local codling moth populations. Nachr. Dtsch. Pflanzenschutzd. 2005, 57, 29–34. [Google Scholar]
- Sauphanor, B.; Berling, M.; Toubon, J.-F.; Reyes, M.; Delnatte, J. Carpocapse des pommes cas de résistance aux virus de la granulose dans le Sud-Est. Phytoma 2006, 590, 24–27. [Google Scholar]
- Graillot, B.; Berling, M.; Blachere-López, C.; Siegwart, M.; Besse, S.; López-Ferber, M. Progressive adaptation of a CpGV isolate to codling moth populations resistant to CpGV-M. Viruses 2014, 6, 5135–5144. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Wennmann, J.T.; Wang, D.; Jehle, J.A. Single nucleotide polymorphism (SNP) frequencies and distribution reveal complex genetic composition of seven novel natural isolates of Cydia pomonella granulovirus. Virology 2020, 541, 32–40. [Google Scholar] [CrossRef]
- Gueli-Alletti, G.; Sauer, A.J.; Weihrauch, B.; Fristch, E.; Undorf-Spahn, K.; Wennmann, J.T.; Jehle, J.A. Using Next Generation Sequencing to Identify and Quantify the Genetic Composition of Resistance-Breaking Commercial Isolates of Cydia pomonella Granulovirus. Viruses 2017, 9, 250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegwart, M.; Mauguin, S.; Besse, S.; Lopez-Ferber, M.; Hinsberger, A.; Gauffre, B. Le carpocapse des pommes résiste au virus de la granulose. Phytoma St. Végétaux 2020, 738, 45–50. [Google Scholar]
- Asser-Kaiser, S.; Radtke, P.; El-Salamouny, S.; Winstanley, D.; Jehle, J.A. Baculovirus resistance in codling moth (Cydia pomonella L.) caused by early block of virus replication. Virology 2011, 410, 360–367. [Google Scholar] [CrossRef] [Green Version]
- Graillot, B.; Bayle, S.; Blachere-Lopez, C.; Besse, S.; Siegwart, M.; Lopez-Ferber, M. Biological characteristics of experimental genotype mixtures of Cydia pomonella granulovirus (CpGV): Ability to control susceptible and resistant pest populations. Viruses 2016, 8, 147. [Google Scholar] [CrossRef]
- Marsberg, T.; Jukes, M.D.; Krejmer-Rabalska, M.; Rabalski, L.; Knox, C.M.; Moore, S.D.; Hill, M.P.; Szewczyk, B. Morphological, genetic and biological characterisation of a novel Alphabaculovirus isolated from Cryptophlebia peltastica (Lepidoptera: Tortricidae). J. Invertebr. Pathol. 2018, 157, 90–99. [Google Scholar] [CrossRef]
- Wennmann, J.T.; Eigenbrod, M.; Marsberg, T.; Moore, S.D.; Knox, C.M.; Hill, M.P.; Jehle, J.A. Cryptophlebia peltastica nucleopolyhedrovirus is highly infectious to codling moth larvae and cells. Appl. Environ. Microbiol. 2019, 85, e00795-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berling, M.; Blachere-Lopez, C.; Soubabere, O.; Lery, X.; Bonhomme, A.; Sauphanor, B.; Lopez-Ferber, M. Cydia pomonella granulovirus genotypes overcome virus resistance in the codling moth and improve virus efficiency by selection against resistant hosts. Appl. Environ. Microbiol. 2009, 75, 925–930. [Google Scholar] [CrossRef] [Green Version]
- Tanada, Y. A granulosis virus of the codling moth, Carpocapsa [Cydia] pomonella (Linnaeus) (Olethreutidae, Lepidoptera) [from Mexico]. J. Insect Pathol. 1964, 6, 378. [Google Scholar]
- Luque, T.; Finch, R.; Crook, N.; O’Reilly, D.R.; Winstanley, D. The complete sequence of the Cydia pomonella granulovirus genome. J. Gen. Virol. 2001, 82, 2531–2547. [Google Scholar] [CrossRef]
- Graillot, B.; Besse, S.; Blachère-Lopez, C.; Olivares, J.; Graillot, B.; Olivares, J.; Siegwart, M.; López-Ferber, M. Sequence analysis of CpGV-R5 isolate, able to efficiently control CpGV-M resistant insects: Relation between biologic al activity and genome. IOBC-WPRS Bull. 2013, 90, 195–199. [Google Scholar]
- Graillot, B.; Berling, M.; Blachere-López, C.; Siegwart, M.; Besse, S.; López-Ferber, M. Correction: Progressive adaptation of a CpGV isolate to codling moth populations resistant to CpGV-M. Viruses 2015, 7, 6313–6315. [Google Scholar] [CrossRef] [Green Version]
- Hinsberger, A.; Theulier Saint Germain, S.; Guerrero, P.; Blachère-López, C.; López-Ferber, M.; Bayle, S. A combination of real time PCR and high resolution melting analysis to detect and identify CpGV genotypes involved in type I resistance. Viruses 2019, 11, 723. [Google Scholar] [CrossRef] [Green Version]
- Hinsberger, A.; Blachère-Lopez, C.; Lopez-Ferber, M. Promoting mixed genotype infections in CpGV: Analysis on field and laboratory sprayed apple leaves. Biocontrol Sci. Technol. 2020, 30, 975–982. [Google Scholar] [CrossRef]
- Gebhardt, M.M.; Eberle, K.E.; Radtke, P.; Jehle, J.A. Baculovirus resistance in codling moth is virus isolate-dependent and the consequence of a mutation in viral gene pe38. Proc. Natl. Acad. Sci. USA 2014, 111, 15711–15716. [Google Scholar] [CrossRef] [Green Version]
- Tanada, Y. Synergism between two viruses of the armyworm, Pseudaletia unipuncta (Haworth) (Lepidoptera: Noctuidae). J. Insect Pathol. 1959, 1, 215–231. [Google Scholar]
- Tanada, Y. A synopsis of studies on the synergistic property of an insect baculovirus: A tribute to Edward A. Steinhaus. J. Invertebr. Pathol. 1985, 45, 125–138. [Google Scholar] [CrossRef]
- Derksen, A.C.G.; Granados, R.R. Alteration of a lepidopteran peritrophic membrane by baculoviruses and enhancement of viral infectivity. Virology 1988, 167, 242–250. [Google Scholar] [CrossRef]
- Roelvink, P.W.; Corsaro, B.G.; Granados, R.R. Characterization of the Helicoverpa armigera and Pseudaletia unipuncta granulovirus enhancin genes. J. Gen. Virol. 1995, 76, 2693–2705. [Google Scholar] [CrossRef] [PubMed]
- Tanada, Y.; Hukuhara, T. Enhanced infection of a nuclear-polyhedrosis virus in larvae of the armyworm, Pseudaletia unipuncta, by a factor in the capsule of a granulosis virus. J. Invertebr. Pathol. 1971, 17, 116–126. [Google Scholar] [CrossRef]
- Kikhno, I.; Gutiérrez, S.; Croizier, L.; Croizier, G.; López-Ferber, M. Characterization of pif, a gene required for the per os infectivity of Spodoptera littoralis nucleopolyhedrovirus. J. Gen. Virol. 2002, 83, 3013–3022. [Google Scholar] [CrossRef]
- Flipsen, J.T.M.; Martens, J.W.M.; Van Oers, M.M.; Vlak, J.M.; Van Lent, J.W.M. Passage of Autographa californica nuclear polyhedrosis virus through the midgut epithelium of Spodoptera exigua larvae. Virology 1995, 208, 328–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, H.; Miyagawa, M. Regeneration of midgut epithelial cell in the silkworm, Bombyx mori, infected with viruses. J. Invertebr. Pathol. 1978, 32, 373–380. [Google Scholar] [CrossRef]
- Levy, S.M.; Falleiros, A.M.F.; Gregório, E.A.; Arrebola, N.R.; Toledo, L.A. The larval midgut of Anticarsia gemmatalis (Hübner) (Lepidoptera: Noctuidae): Light and electron microscopy studies of the epithelial cells. Braz. J. Biol. 2004, 64, 633–638. [Google Scholar] [CrossRef] [Green Version]
- Beperet, I.; Irons, S.L.; Simón, O.; King, L.A.; Williams, T.; Possee, R.D.; Lopez-Ferber, M.; Caballero, P. Superinfection exclusion in Alphabaculovirus infections is concomitant with actin reorganization. J. Virol. 2014, 88, 3548–3556. [Google Scholar] [CrossRef] [Green Version]
- Volkman, L.E. Baculovirus infectivity and the actin cytoskeleton. Curr. Drug Targets 2007, 8, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Hinsberger, A.; Graillot, B.; Blachere-Lopez, C.; Juliant, S.; Cerutti, M.; King, L.A.; Possee, R.D.; Gallardo, F.; Lopez-Ferber, M. Tracing Baculovirus AcMNPV Infection Using a Real-Time Method Based on ANCHORTM DNA Labeling Technology. Viruses 2020, 12, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Virus Replication | |||||||
|---|---|---|---|---|---|---|---|
| Host | Virus Inoculum | CpGV-M | CpGV-R5 | CrpeNPV | CpGV-M + CrpeNPV | CpGV-R5 + CrpeNPV | Total Number of Larvae |
| CpNPP | CpGV-M | 11 | - | - | - | - | 12 |
| CpGV-R5 | - | 8 | - | - | - | 12 | |
| CpGV-M + CrpeNPV | 1 | - | 2 | 11 | - | 15 | |
| CpGV-R5 + CrpeNPV | - | 2 | 2 | - | 10 | 15 | |
| RGV | CpGV-M | 0 | - | - | - | - | 13 |
| CpGV-R5 | - | 20 | - | - | - | 25 | |
| CpGV-M + CrpeNPV | 0 | - | 19 | 0 | - | 30 | |
| CpGV-R5 + CrpeNPV | - | 7 | 1 | - | 7 | 22 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinsberger, A.; Blachère-Lopez, C.; Knox, C.; Moore, S.; Marsberg, T.; Lopez-Ferber, M. CpGV-M Replication in Type I Resistant Insects: Helper Virus and Order of Ingestion Are Important. Viruses 2021, 13, 1695. https://doi.org/10.3390/v13091695
Hinsberger A, Blachère-Lopez C, Knox C, Moore S, Marsberg T, Lopez-Ferber M. CpGV-M Replication in Type I Resistant Insects: Helper Virus and Order of Ingestion Are Important. Viruses. 2021; 13(9):1695. https://doi.org/10.3390/v13091695
Chicago/Turabian StyleHinsberger, Aurélie, Christine Blachère-Lopez, Caroline Knox, Sean Moore, Tamryn Marsberg, and Miguel Lopez-Ferber. 2021. "CpGV-M Replication in Type I Resistant Insects: Helper Virus and Order of Ingestion Are Important" Viruses 13, no. 9: 1695. https://doi.org/10.3390/v13091695






