Successful Clearance of 300 Day SARS-CoV-2 Infection in a Subject with B-Cell Depletion Associated Prolonged (B-DEAP) COVID by REGEN-COV Anti-Spike Monoclonal Antibody Cocktail
Abstract
:1. Introduction
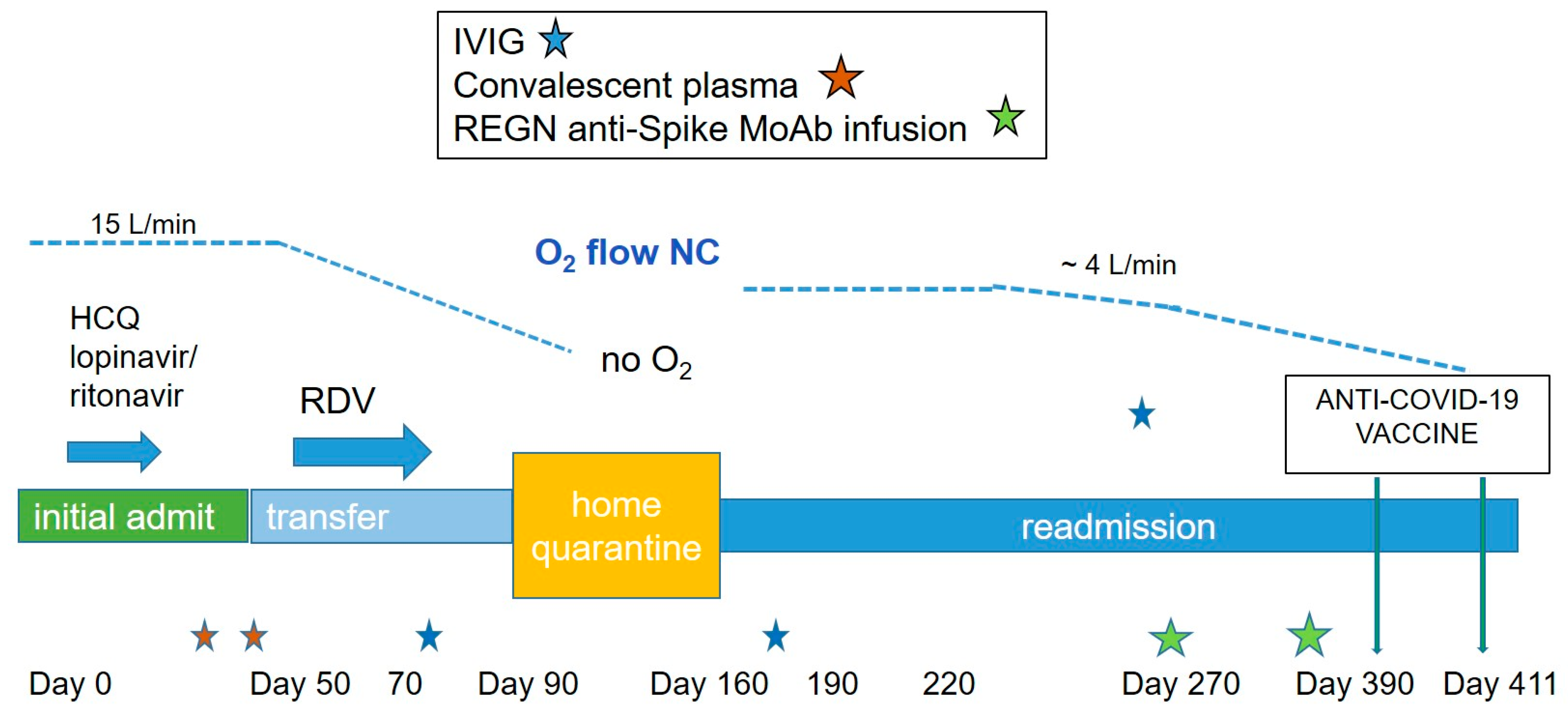
Case Report Presentation
2. Materials and Methods
2.1. Sample Processing Protocols
2.2. Serology Tests
2.2.1. ReSARS® CoV-2 N-Protein and S-RBD IgG ELISA Kits for RUO, Zalgen Labs, LLC
2.2.2. SARS-CoV-2 Anti-S (HEXAPRO) ELISA for RUO (Dr Robinson’s Lab)
2.2.3. COVID-19 ACE2 Competition Assay® (V-PLEX SARS-CoV-2 Panel 2), Meso Scale Discovery, for RUO
2.3. Molecular Tests
2.3.1. CDC Emergency Use Authorization (EUA) 2019-nCoV Real-Time qRT-PCR Panel Protocol, A. Smither and Dr. Garry’s Lab. for RUO
2.3.2. Applied Biosystems TaqPath COVID-19 Combo Kit (Dr. Tian’s Lab) for IVD Use
2.3.3. SARS-CoV-2 Genome Sequencing (Andersen Lab. Scripps Research Institute), for RUO
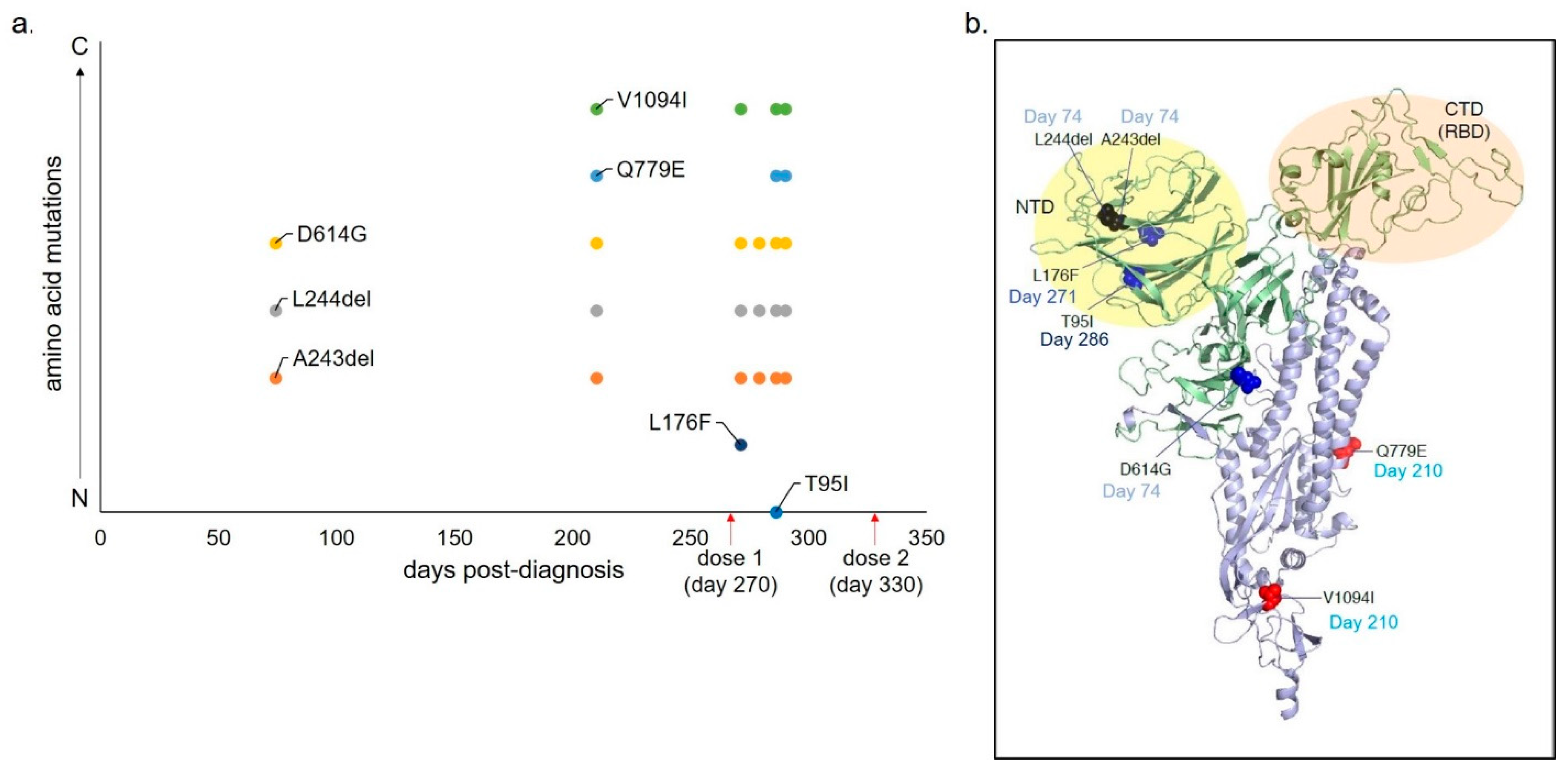
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choi, B.; Choudhary, M.C.; Regan, J.; Sparks, J.A.; Padera, R.F.; Qiu, X.; Solomon, I.H.; Kuo, H.-H.; Boucau, J.; Bowman, K.; et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020, 383, 2291–2293. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Dan, J.M.; Mateus, J.; Kato, Y.; Hastie, K.M.; Yu, E.D.; Faliti, C.E.; Grifoni, A.; Ramirez, S.I.; Haupt, S.; Frazier, A.; et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 2021, 371. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.E.; Hastie, K.M.; Cross, R.W.; Yenni, R.E.; Elliott, D.H.; Rouelle, J.A.; Kannadka, C.B.; Smira, A.A.; Garry, C.E.; Bradley, B.T.; et al. Most neutralizing human monoclonal antibodies target novel epitopes requiring both Lassa virus glycoprotein subunits. Nat. Commun. 2016, 7, 11544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinreich, D.M.; Sivapalasingam, S.; Perry, C.; Pan, C.; Hosain, R.; Mahmood, A.; Davis, J.D.; Turner, K.C.; Hooper, A.T.; Hamilton, J.D.; et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with COVID-19. N. Engl. J. Med. 2021, 384, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Kos, I.; Balensiefer, B.; Roth, S.; Ahlgrimm, M.; Sester, M.; Schmidt, T.; Thurner, L.; Bewarder, M.; Bals, R.; Lammert, F.; et al. Prolonged Course of COVID-19-Associated Pneumonia in a B-Cell Depleted Patient After Rituximab. Front. Oncol. 2020, 10, 1578. [Google Scholar] [CrossRef]
- Baker, D.; Roberts, C.A.K.; Pryce, G.; Kang, A.S.; Marta, M.; Reyes, S.; Schmierer, K.; Giovannoni, G.; Amor, S. COVID-19 vaccine-readiness for anti-CD20-depleting therapy in autoimmune diseases. Clin. Exp. Immunol. 2020, 202, 149–161. [Google Scholar] [CrossRef]
- McLaughlin, P.; Grillo-López, A.J.; Link, B.K.; Levy, R.; Czuczman, M.S.; Williams, M.E.; Heyman, M.R.; Bence-Bruckler, I.; White, C.A.; Cabanillas, F.; et al. Rituximab chimeric anti-CD20 monoclonal antibody therapy for relapsed indolent lymphoma: Half of patients respond to a four-dose treatment program. J. Clin. Oncol. 1998, 16, 2825–2833. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Calle, N.; Hartley, S.; Ahearne, M.; Kasenda, B.; Beech, A.; Knight, H.; Balotis, C.; Kennedy, B.; Wagner, S.; Dyer, M.; et al. Kinetics of T-cell subset reconstitution following treatment with bendamustine and rituximab for low-grade lymphoproliferative disease: A population-based analysis. Br. J. Haematol. 2018, 184, 957–968. [Google Scholar] [CrossRef]
- Lamure, S.; Dulery, R.; Demord, M.; Di Blasi, R.; Chauchet, A.; Hueso, T.; Rossi, C.; Drenou, B.; Deau-Fischer, B.; Sousain, C.; et al. High incidence of persistent COVID-19 among patients with lymphoma treated with B-cell depleting immunotherapy. AACR 2021, S09-02. [Google Scholar] [CrossRef]
- Yasuda, H.; Tsukune, Y.; Watanabe, N.; Sugimoto, K.; Uchimura, A.; Tateyama, M.; Miyashita, Y.; Ochi, Y.; Komatsu, N. Persistent COVID-19 pneumonia and failure to develop Anti-SARS-CoV-2 antibodies during Rituximab maintenance therapy for follicular lymphoma. Clin. Lymphoma Myeloma Leuk. 2020, 20, 774–776. [Google Scholar] [CrossRef] [PubMed]
- Veeramani, S.; Wang, S.-Y.; Dahle, C.; Blackwell, S.; Jacobus, L.; Knutson, T.; Button, A.; Link, B.K.; Weiner, G.J. Rituximab infusion induces NK activation in lymphoma patients with the high-affinity CD16 polymorphism. Blood 2011, 118, 3347–3349. [Google Scholar] [CrossRef] [Green Version]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M.; et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 2020, 369, 1014–1018. [Google Scholar] [CrossRef] [PubMed]
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; Ferreira, I.A.T.M.; Gayed, S.; Jahun, A.; Hosmillo, M.; Rees-Spear, C.; Mlcochova, P.; Lumb, I.U.; et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021, 592, 277–282. [Google Scholar] [CrossRef]
- Irie, E.; Shirota, Y.; Suzuki, C.; Tajima, Y.; Ishizawa, K.; Kameoka, J.; Harigae, H.; Ishii, T. Severe hypogammaglobulinemia persisting for 6 years after treatment with rituximab combined chemotherapy due to arrest of B lymphocyte differentiation together with alteration of T lymphocyte homeostasis. Int. J. Hematol. 2010, 91, 501–508. [Google Scholar] [CrossRef]
- Sakai, H.; Tanaka, Y.; Tazawa, H.; Shimizu, S.; Verma, S.; Ohira, M.; Tahara, H.; Ide, K.; Ishiyama, K.; Kobayashi, T.; et al. Effect of Fc-gamma Receptor Polymorphism on Rituximab-Mediated B Cell Depletion in ABO-Incompatible Adult Living Donor Liver Transplantation. Transpl. Direct 2017, 3, e164. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.; Tabchi, S.; Hagemeister, F. Obinutuzumab Treatment of Follicular Lymphoma. N. Engl. J. Med. 2017, 377, 2605–2606. [Google Scholar] [CrossRef] [PubMed]
- Grigg, A.; Dyer, M.J.; Díaz, M.G.; Dreyling, M.; Rule, S.; Lei, G.; Knapp, A.; Wassner-Fritsch, E.; Marlton, P. Safety and efficacy of obinutuzumab with CHOP or bendamustine in previously untreated follicular lymphoma. Haematologica 2016, 102, 765–772. [Google Scholar] [CrossRef] [Green Version]
- Niscola, P.; Palombi, M.; Fratoni, S.; Trawinska, M.M.; Scaramucci, L.; Giovannini, M.; Perrotti, A.; De Fabritiis, P. Unusual sequence of lymphoid disorders: Follicular lymphoma subsequent to Hodgkin lymphoma and transformed into diffuse large B-cells non Hodgkin lymphoma. Acta Oncol. 2009, 48, 1073–1074. [Google Scholar] [CrossRef] [PubMed]
- Gratzinger, D.; Jaffe, E.S.; Chadburn, A.; Chan, J.K.C.; de Jong, D.; Goodland, J.R.; Said, J.; Natkunam, Y. Primary/Congenital Immunodeficiency: 2015 SH/EAHP Workshop Report-Part 5. Am. J. Clin. Pathol. 2017, 147, 204–216. [Google Scholar] [CrossRef] [PubMed]

| Primer/Probe Mix | Sequence | |
|---|---|---|
| 2019-nCoV_N1 | Forward | 5′-GACCCCAAAATCAGCGAAAT-3′ |
| Reverse | 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ | |
| Probe | 5′-FAM-ACCCCGCAT/ZEN/TACGTTTGGTGGACC-3IABkFQ-3′ | |
| 2019-nCoV_N2 | Forward | 5′-TTACAAACATTGGCCGCAAA-3′ |
| Reverse | 5′-GCGCGACATTCCGAAGAA-3′ | |
| Probe | 5′-FAM-ACAATTTGC/ZEN/CCCCAGCGCTTCAG-3IABkFQ-3′ | |
| Hu RNAseP (RP) | Forward | 5′-AGATTTGGACCTGCGAGCG-3′ |
| Reverse | 5′-GAGCGGCTGTCTCCACAAGT-3′ | |
| Probe | 5′-FAM-TTC TGA CCT/ZEN/GAA GGC TCT GCG CG-3IABkFQ-3′ | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drouin, A.C.; Theberge, M.W.; Liu, S.Y.; Smither, A.R.; Flaherty, S.M.; Zeller, M.; Geba, G.P.; Reynaud, P.; Rothwell, W.B.; Luk, A.P.; et al. Successful Clearance of 300 Day SARS-CoV-2 Infection in a Subject with B-Cell Depletion Associated Prolonged (B-DEAP) COVID by REGEN-COV Anti-Spike Monoclonal Antibody Cocktail. Viruses 2021, 13, 1202. https://doi.org/10.3390/v13071202
Drouin AC, Theberge MW, Liu SY, Smither AR, Flaherty SM, Zeller M, Geba GP, Reynaud P, Rothwell WB, Luk AP, et al. Successful Clearance of 300 Day SARS-CoV-2 Infection in a Subject with B-Cell Depletion Associated Prolonged (B-DEAP) COVID by REGEN-COV Anti-Spike Monoclonal Antibody Cocktail. Viruses. 2021; 13(7):1202. https://doi.org/10.3390/v13071202
Chicago/Turabian StyleDrouin, Arnaud C., Marc W. Theberge, Sharon Y. Liu, Allison R. Smither, Shelby M. Flaherty, Mark Zeller, Gregory P. Geba, Peter Reynaud, W. Benjamin Rothwell, Alfred P. Luk, and et al. 2021. "Successful Clearance of 300 Day SARS-CoV-2 Infection in a Subject with B-Cell Depletion Associated Prolonged (B-DEAP) COVID by REGEN-COV Anti-Spike Monoclonal Antibody Cocktail" Viruses 13, no. 7: 1202. https://doi.org/10.3390/v13071202






