HIV-1 Packaging Visualised by In-Gel SHAPE
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Preparation
2.2. Protein Expression and Purification
2.3. In-Gel SHAPE
2.4. Electrophoresis Mobility Shift Assay (EMSA)
2.5. Reverse Transcription, Sequencing and Data Analysis
2.6. Cross-Linking
2.7. Statistical Analysis
3. Results
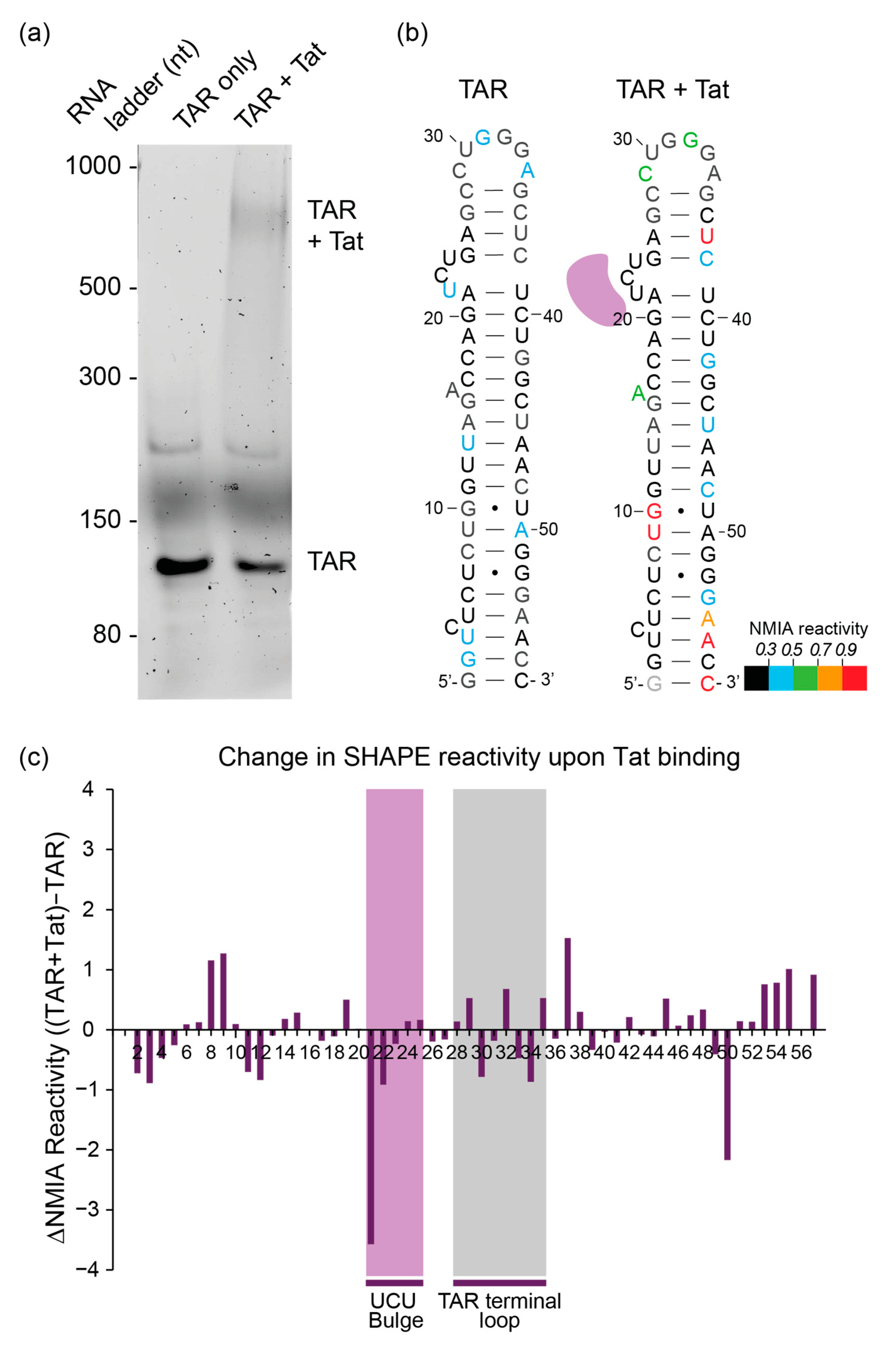
3.1. In-Gel SHAPE of an RNA–Protein Complex Accurately Reports upon Its RNA Structure
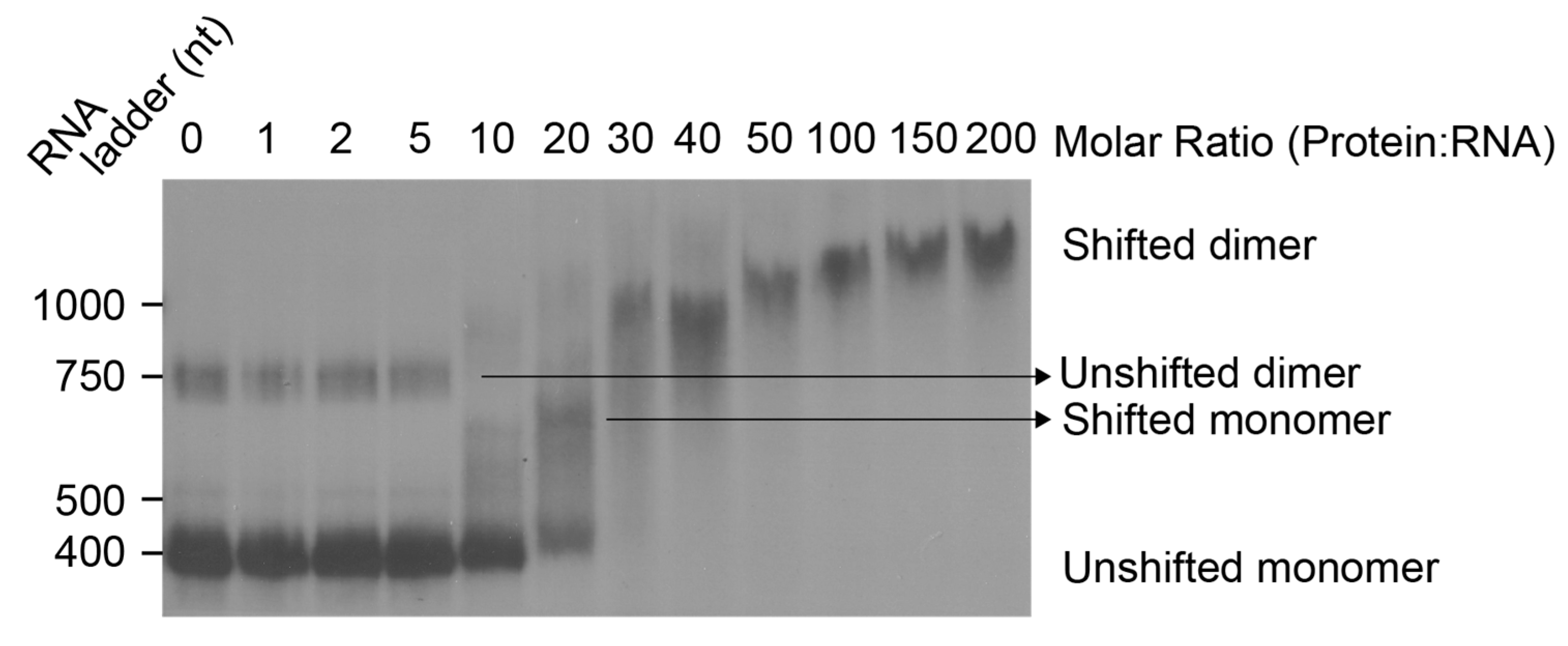
3.2. Initial Contacts between Gag Protein and HIV-1 Leader RNA Shift the RNA Structural Equilibrium
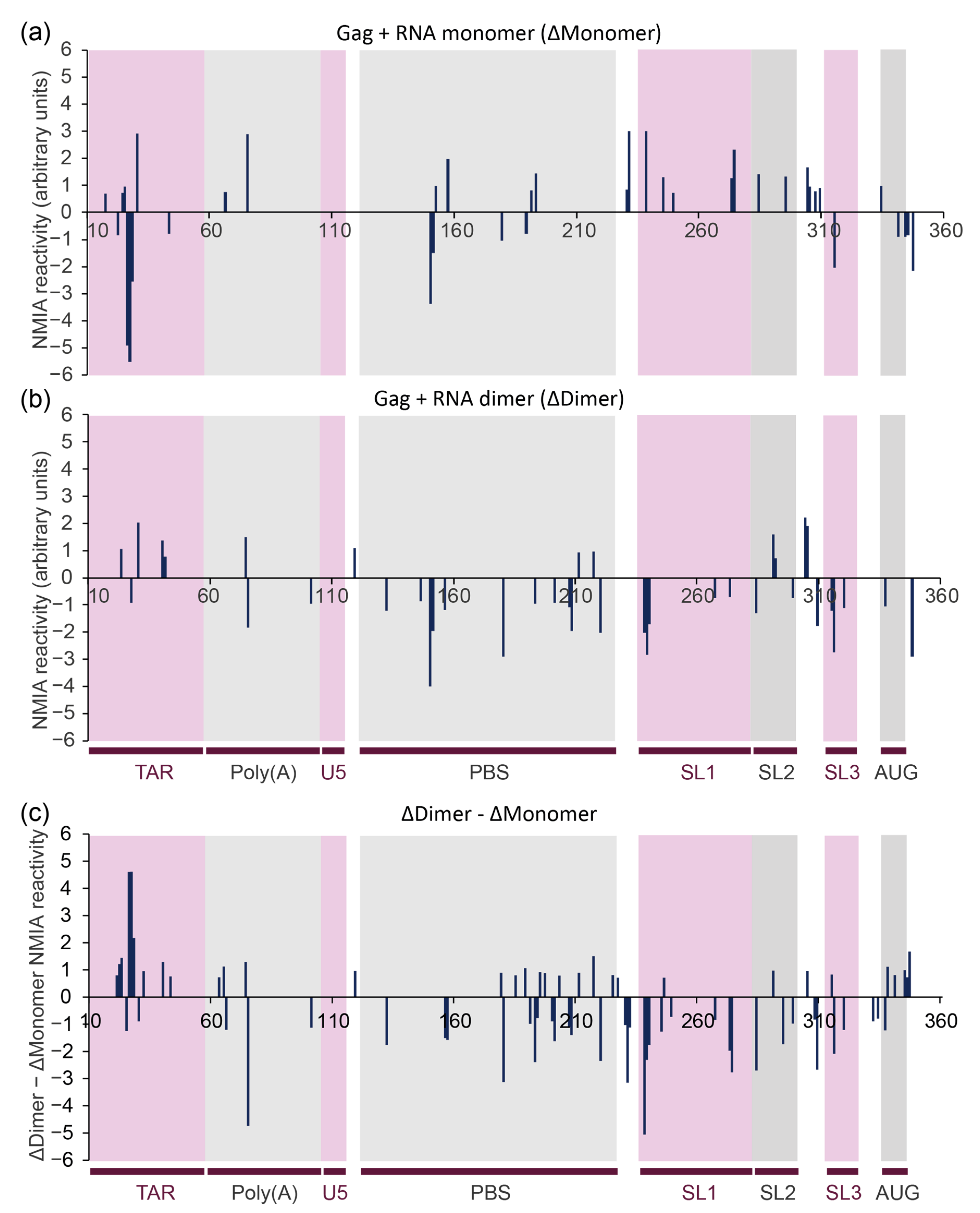
3.3. In-Gel SHAPE Suggests That Gag Remodels the Monomeric RNA Conformation but Stabilises the Dimer
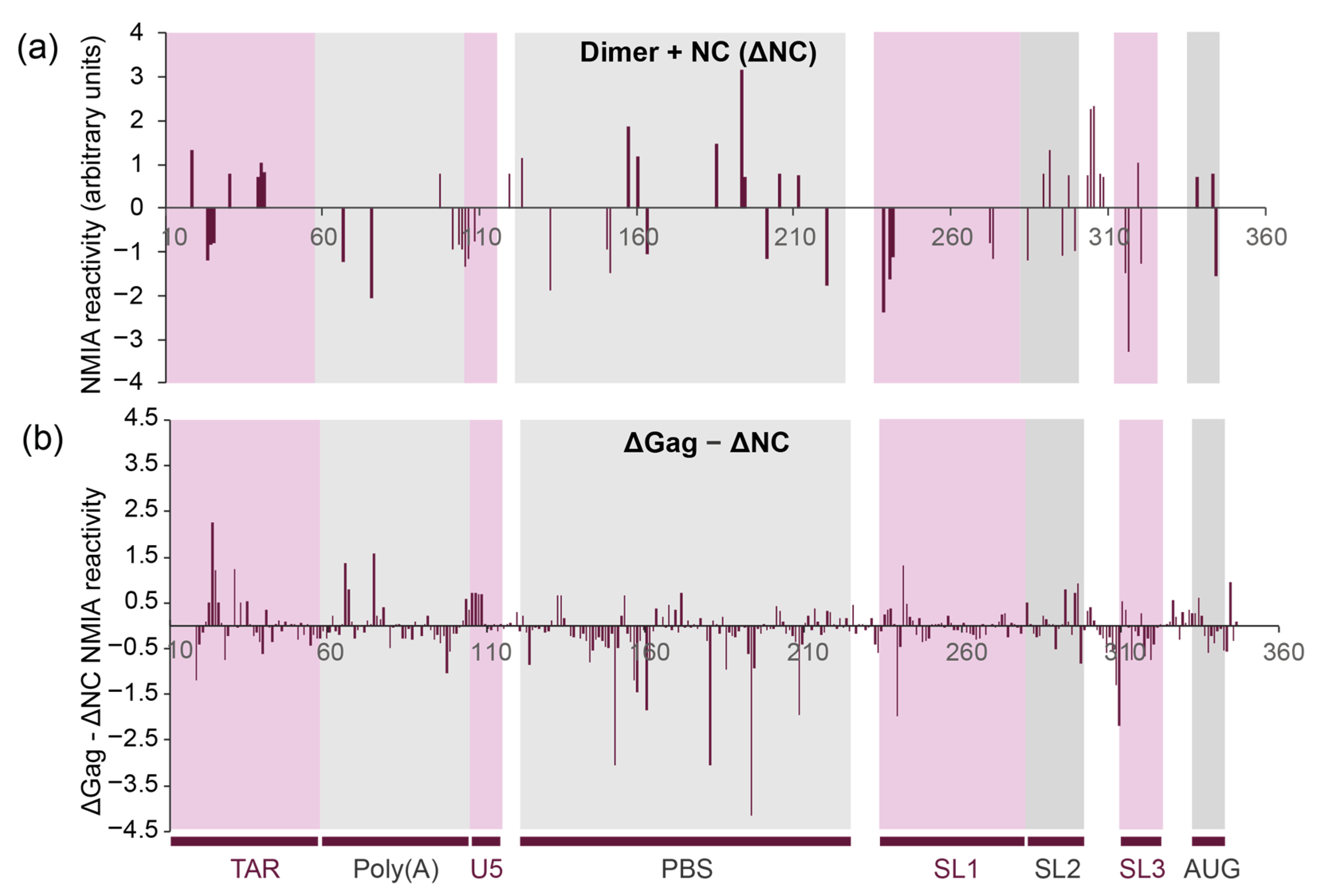
3.4. NC and Gag Have Different Effects upon the RNA Structure, Commensurate with Their Roles in the Viral Lifecycle
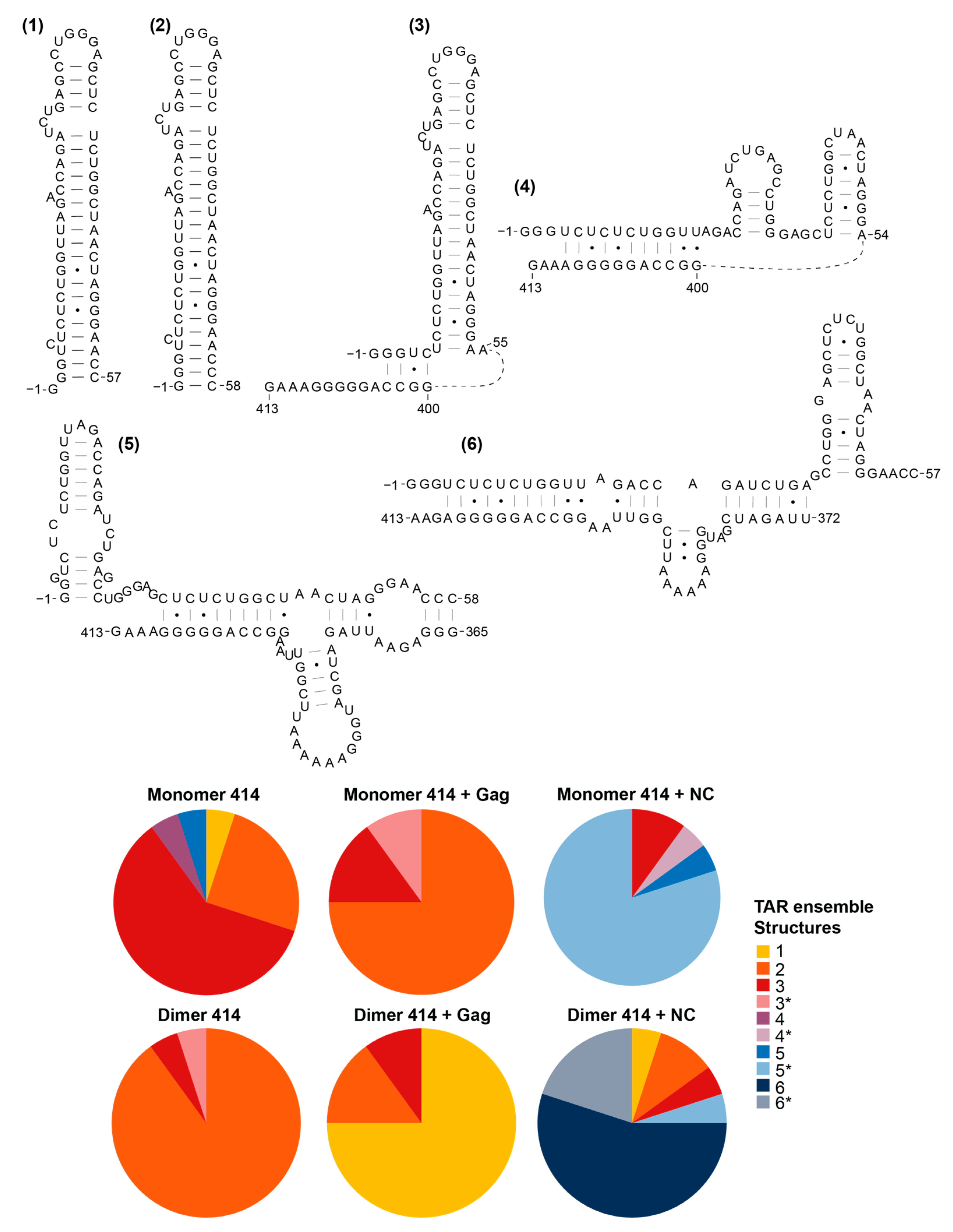
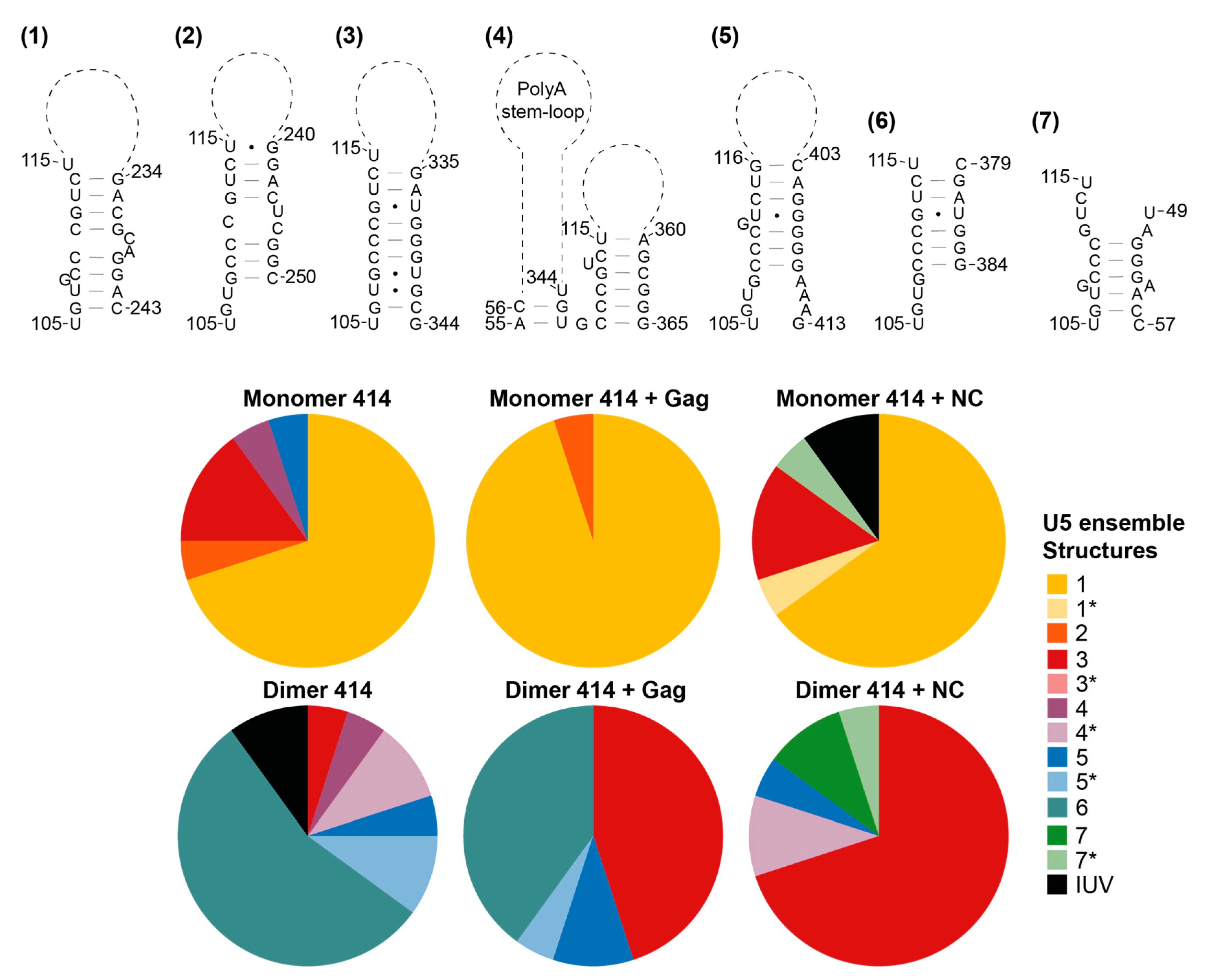
3.5. The Structural Ensembles of the RNA Change upon Dimerisation or Ligand Binding, and Differ between NC-Bound and Gag-Bound RNA
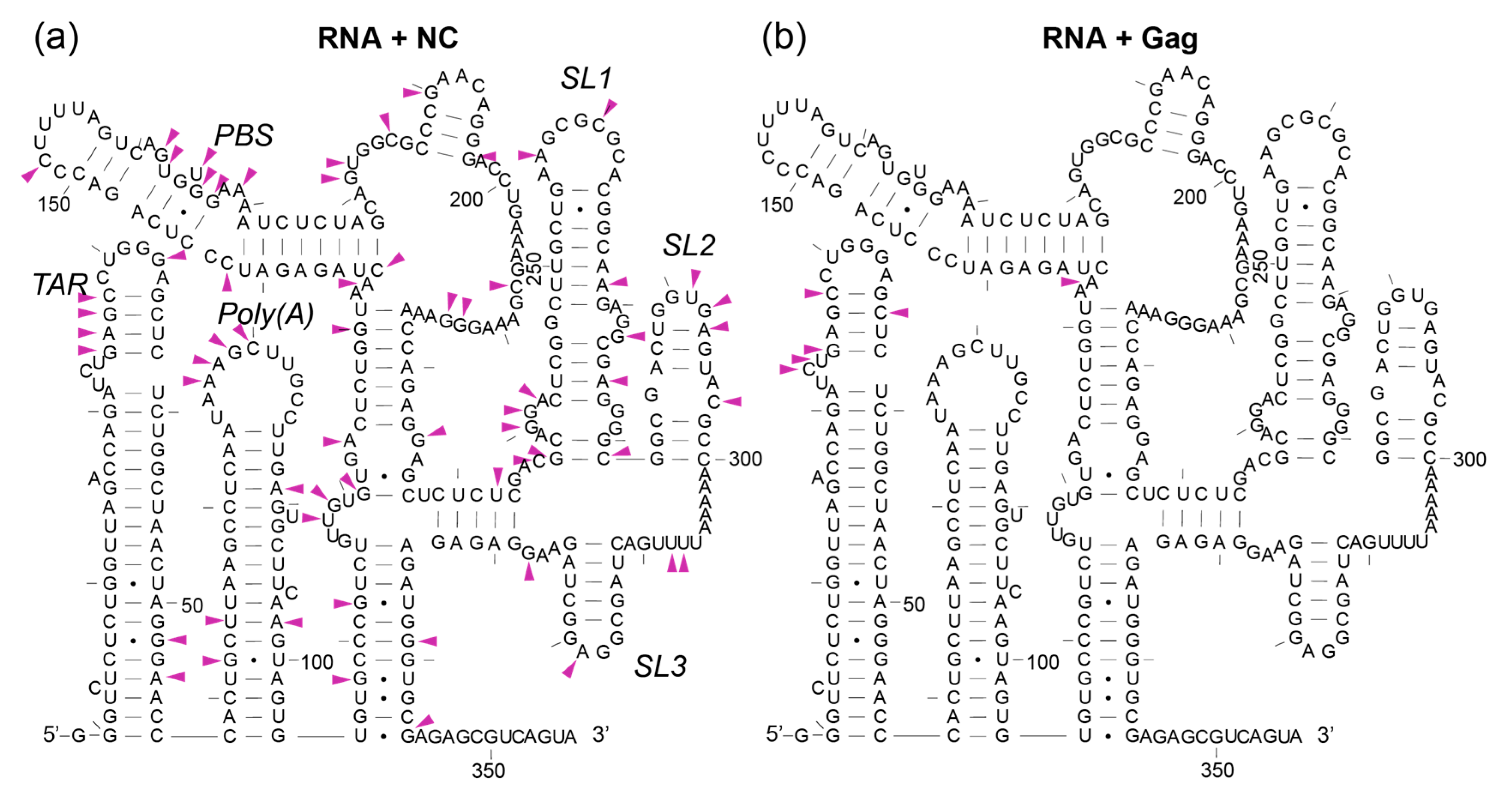
3.6. The First Gag Binding Sites Are in TAR and Poly(A), Whereas NC Reacts Extensively at Multiple Sites
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Freed, E.O. HIV-1 Gag Proteins: Diverse Functions in the Virus Life Cycle. Virology 1998, 251, 1–15. [Google Scholar] [CrossRef]
- Rodriguez-Rodriguez, L.; Tsuchihashi, Z.; Fuentes, G.M.; Bambara, R.A.; Fay, P.J. Influence of Human Immunodeficiency Virus Nucleocapsid Protein on Synthesis and Strand Transfer by the Reverse Transcriptase In Vitro. J. Biol. Chem. 1995, 270, 15005–15011. [Google Scholar] [CrossRef]
- Huang, Y.; Khorchid, A.; Gabor, J.; Wang, J.; Li, X.; Darlix, J.-L.; Wainberg, M.A.; Kleiman, L. The Role of Nucleocapsid and U5 Stem/A-Rich Loop Sequences in TRNA3Lys Genomic Placement and Initiation of Reverse Transcription in Human Immunodeficiency Virus Type 1. J. Virol. 1998, 72, 3907–3915. [Google Scholar] [CrossRef]
- Cen, S.; Huang, Y.; Khorchid, A.; Darlix, J.-L.; Wainberg, M.A.; Kleiman, L. The Role of Pr55gag in the Annealing of TRNA3Lys to Human Immunodeficiency Virus Type 1 Genomic RNA. J. Virol. 1999, 73, 4485–4488. [Google Scholar] [CrossRef]
- Feng, Y.-X.; Campbell, S.; Harvin, D.; Ehresmann, B.; Ehresmann, C.; Rein, A. The Human Immunodeficiency Virus Type 1 Gag Polyprotein Has Nucleic Acid Chaperone Activity: Possible Role in Dimerization of Genomic RNA and Placement of TRNA on the Primer Binding Site. J. Virol. 1999, 73, 4251–4256. [Google Scholar] [CrossRef]
- Hargittai, M.R.S.; Mangla, A.T.; Gorelick, R.J.; Musier-Forsyth, K. HIV-1 Nucleocapsid Protein Zinc Finger Structures Induce TRNALys, 3 Structural Changes but Are Not Critical for Primer/Template Annealing11Edited by M. F. Summers. J. Mol. Biol. 2001, 312, 985–997. [Google Scholar] [CrossRef]
- Hargittai, M.R.S.; Gorelick, R.J.; Rouzina, I.; Musier-Forsyth, K. Mechanistic Insights into the Kinetics of HIV-1 Nucleocapsid Protein-Facilitated TRNA Annealing to the Primer Binding Site. J. Mol. Biol. 2004, 337, 951–968. [Google Scholar] [CrossRef]
- Rong, L.; Liang, C.; Hsu, M.; Guo, X.; Roques, B.P.; Wainberg, M.A. HIV-1 Nucleocapsid Protein and the Secondary Structure of the Binary Complex Formed between TRNALys.3 and Viral RNA Template Play Different Roles during Initiation of (−) Strand DNA Reverse Transcription. J. Biol. Chem. 2001, 276, 47725–47732. [Google Scholar] [CrossRef]
- Ingemarsdotter, C.K.; Zeng, J.; Long, Z.; Lever, A.M.L.; Kenyon, J.C. An RNA-Binding Compound That Stabilizes the HIV-1 GRNA Packaging Signal Structure and Specifically Blocks HIV-1 RNA Encapsidation. Retrovirology 2018, 15, 25. [Google Scholar] [CrossRef]
- Sakuragi, J.; Shioda, T.; Panganiban, A.T. Duplication of the Primary Encapsidation and Dimer Linkage Region of Human Immunodeficiency Virus Type 1 RNA Results in the Appearance of Monomeric RNA in Virions. J. Virol. 2001, 75, 2557–2565. [Google Scholar] [CrossRef]
- Nikolaitchik, O.A.; Dilley, K.A.; Fu, W.; Gorelick, R.J.; Tai, S.-H.S.; Soheilian, F.; Ptak, R.G.; Nagashima, K.; Pathak, V.K.; Hu, W.-S. Dimeric RNA Recognition Regulates HIV-1 Genome Packaging. PLoS Pathog. 2013, 9, e1003249. [Google Scholar] [CrossRef]
- Kuzembayeva, M.; Dilley, K.; Sardo, L.; Hu, W.-S. Life of Psi: How Full-Length HIV-1 RNAs Become Packaged Genomes in the Viral Particles. Virology 2014, 454–455, 362–370. [Google Scholar] [CrossRef]
- Rein, A. RNA Packaging in HIV. Trends Microbiol. 2019, 27, 715–723. [Google Scholar] [CrossRef]
- Berkhout, B. Structure and Function of the Human Immunodeficiency Virus Leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996, 54, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.L.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A.; et al. NMR Detection of Structures in the HIV-1 5′-Leader RNA That Regulate Genome Packaging. Science 2011, 334, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; Ooms, M.; Beerens, N.; Huthoff, H.; Southern, E.; Verhoef, K. In Vitro Evidence That the Untranslated Leader of the HIV-1 Genome Is an RNA Checkpoint That Regulates Multiple Functions through Conformational Changes. J. Biol. Chem. 2002, 277, 19967–19975. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.C.; Marquet, R.; Skripkin, E.; Ehresmann, B.; Ehresmann, C. Mutational Analysis of the Bipartite Dimer Linkage Structure of Human Immunodeficiency Virus Type 1 Genomic RNA. J. Biol. Chem. 1994, 269, 27486–27493. [Google Scholar] [CrossRef]
- Paillart, J.; Marquet, R.; Skripkin, E.; Ehresmann, C.; Ehresmann, B. Dimerization of Retroviral Genomic RNAs: Structural and Functional Implications. Biochimie 1996, 78, 639–653. [Google Scholar] [CrossRef]
- Paillart, J.C.; Skripkin, E.; Ehresmann, B.; Ehresmann, C.; Marquet, R. A Loop-Loop “Kissing” Complex Is the Essential Part of the Dimer Linkage of Genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 1996, 93, 5572–5577. [Google Scholar] [CrossRef]
- Paillairt, J.-C.; Skripkin, E.; Ehresmann, B.; Ehresmann, C.; Marquet, R. The Use of Chemical Modification Interference and Inverse PCR Mutagenesis to Identify the Dimerization Initiation Site of HIV-1 Genomic RNA. Pharm. Acta Helv. 1996, 71, 21–28. [Google Scholar] [CrossRef]
- Skripkin, E.; Paillart, J.C.; Marquet, R.; Ehresmann, B.; Ehresmann, C. Identification of the Primary Site of the Human Immunodeficiency Virus Type 1 RNA Dimerization In Vitro. Proc. Natl. Acad. Sci. USA 1994, 91, 4945–4949. [Google Scholar] [CrossRef]
- Van Bel, N.; Das, A.T.; Cornelissen, M.; Abbink, T.E.M.; Berkhout, B. A Short Sequence Motif in the 5′ Leader of the HIV-1 Genome Modulates Extended RNA Dimer Formation and Virus Replication. J. Biol. Chem. 2014, 289, 35061–35074. [Google Scholar] [CrossRef]
- Lever, A.; Gottlinger, H.; Haseltine, W.; Sodroski, J. Identification of a Sequence Required for Efficient Packaging of Human Immunodeficiency Virus Type 1 RNA into Virions. J. Virol. 1989, 63, 4085–4087. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, V.; Summers, M.F. How Retroviruses Select Their Genomes. Nat. Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef]
- Bell, N.M.; L’Hernault, A.; Murat, P.; Richards, J.E.; Lever, A.M.L.; Balasubramanian, S. Targeting RNA-Protein Interactions within the Human Immunodeficiency Virus Type 1 Lifecycle. Biochemistry 2013, 52, 9269–9274. [Google Scholar] [CrossRef] [PubMed]
- Clever, J.; Sassetti, C.; Parslow, T.G. RNA Secondary Structure and Binding Sites for Gag Gene Products in the 5′ Packaging Signal of Human Immunodeficiency Virus Type 1. J. Virol. 1995, 69, 2101–2109. [Google Scholar] [CrossRef]
- Abbink, T.E.M.; Berkhout, B. A Novel Long Distance Base-Pairing Interaction in Human Immunodeficiency Virus Type 1 RNA Occludes the Gag Start Codon. J. Biol. Chem. 2003, 278, 11601–11611. [Google Scholar] [CrossRef] [PubMed]
- Mailler, E.; Bernacchi, S.; Marquet, R.; Paillart, J.-C.; Vivet-Boudou, V.; Smyth, R.P. The Life-Cycle of the HIV-1 Gag–RNA Complex. Viruses 2016, 8, 248. [Google Scholar] [CrossRef]
- Huthoff, H.; Berkhout, B. Two Alternating Structures of the HIV-1 Leader RNA. RNA 2001, 7, 143–157. [Google Scholar] [CrossRef]
- Abbink, T.E.M.; Ooms, M.; Haasnoot, P.C.J.; Berkhout, B. The HIV-1 Leader RNA Conformational Switch Regulates RNA Dimerization but Does Not Regulate MRNA Translation. Biochemistry 2005, 44, 9058–9066. [Google Scholar] [CrossRef]
- Kenyon, J.C.; Prestwood, L.J.; Le Grice, S.F.J.; Lever, A.M.L. In-Gel Probing of Individual RNA Conformers within a Mixed Population Reveals a Dimerization Structural Switch in the HIV-1 Leader. Nucleic Acids Res. 2013, 41, e174. [Google Scholar] [CrossRef]
- Masuda, T.; Sato, Y.; Huang, Y.-L.; Koi, S.; Takahata, T.; Hasegawa, A.; Kawai, G.; Kannagi, M. Fate of HIV-1 CDNA Intermediates during Reverse Transcription Is Dictated by Transcription Initiation Site of Virus Genomic RNA. Sci. Rep. 2015, 5, 17680. [Google Scholar] [CrossRef] [PubMed]
- Kharytonchyk, S.; Monti, S.; Smaldino, P.J.; Van, V.; Bolden, N.C.; Brown, J.D.; Russo, E.; Swanson, C.; Shuey, A.; Telesnitsky, A.; et al. Transcriptional Start Site Heterogeneity Modulates the Structure and Function of the HIV-1 Genome. Proc. Natl. Acad. Sci. USA 2016, 113, 13378–13383. [Google Scholar] [CrossRef]
- Brown, J.D.; Kharytonchyk, S.; Chaudry, I.; Iyer, A.S.; Carter, H.; Becker, G.; Desai, Y.; Glang, L.; Choi, S.H.; Singh, K.; et al. Structural Basis for Transcriptional Start Site Control of HIV-1 RNA Fate. Science 2020, 368, 413–417. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Bieniasz, P.D.; Simon, S.M. Imaging the Biogenesis of Individual HIV-1 Virions in Live Cells. Nature 2008, 454, 236–240. [Google Scholar] [CrossRef]
- Fu, W.; Gorelick, R.J.; Rein, A. Characterization of Human Immunodeficiency Virus Type 1 Dimeric RNA from Wild-Type and Protease-Defective Virions. J. Virol. 1994, 68, 5013–5018. [Google Scholar] [CrossRef] [PubMed]
- Jalalirad, M.; Laughrea, M. Formation of Immature and Mature Genomic RNA Dimers in Wild-Type and Protease-Inactive HIV-1: Differential Roles of the Gag Polyprotein, Nucleocapsid Proteins NCp15, NCp9, NCp7, and the Dimerization Initiation Site. Virology 2010, 407, 225–236. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muriaux, D.; Girard, P.-M.; Bonnet-Mathonière, B.; Paoletti, J. Dimerization of HIV-1Lai RNA at Low Ionic Strength an Autocomplementary Sequence in the 5′ Leader Region Is Evidenced by an Antisense Oligonucleotide. J. Biol. Chem. 1995, 270, 8209–8216. [Google Scholar] [CrossRef]
- Laughrea, M.; Jetté, L. A 19-Nucleotide Sequence Upstream of the 5′ Major Splice Donor Is Part of the Dimerization Domain of Human Immunodeficiency Virus 1 Genomic RNA. Biochemistry 1994, 33, 13464–13474. [Google Scholar] [CrossRef]
- Cruceanu, M.; Urbaneja, M.A.; Hixson, C.V.; Johnson, D.G.; Datta, S.A.; Fivash, M.J.; Stephen, A.G.; Fisher, R.J.; Gorelick, R.J.; Casas-Finet, J.R.; et al. Nucleic Acid Binding and Chaperone Properties of HIV-1 Gag and Nucleocapsid Proteins. Nucleic Acids Res. 2006, 34, 593–605. [Google Scholar] [CrossRef]
- Feng, Y.X.; Copeland, T.D.; Henderson, L.E.; Gorelick, R.J.; Bosche, W.J.; Levin, J.G.; Rein, A. HIV-1 Nucleocapsid Protein Induces “Maturation” of Dimeric Retroviral RNA in Vitro. Proc. Natl. Acad. Sci. USA 1996, 93, 7577–7581. [Google Scholar] [CrossRef]
- Ivanchenko, S.; Godinez, W.J.; Lampe, M.; Kräusslich, H.-G.; Eils, R.; Rohr, K.; Bräuchle, C.; Müller, B.; Lamb, D.C. Dynamics of HIV-1 Assembly and Release. PLoS Pathog. 2009, 5, e1000652. [Google Scholar] [CrossRef] [PubMed]
- Kutluay, S.B.; Bieniasz, P.D. Analysis of the Initiating Events in HIV-1 Particle Assembly and Genome Packaging. PLoS Pathog. 2010, 6, e1001200. [Google Scholar] [CrossRef] [PubMed]
- Keane, S.C.; Van, V.; Frank, H.M.; Sciandra, C.A.; McCowin, S.; Santos, J.; Heng, X.; Summers, M.F. NMR Detection of Intermolecular Interaction Sites in the Dimeric 5′-Leader of the HIV-1 Genome. Proc. Natl. Acad. Sci. USA 2016, 113, 13033–13038. [Google Scholar] [CrossRef] [PubMed]
- Beasley, B.E.; Hu, W.-S. Cis-Acting Elements Important for Retroviral RNA Packaging Specificity. J. Virol. 2002, 76, 4950–4960. [Google Scholar] [CrossRef] [PubMed]
- Poole, E.; Strappe, P.; Mok, H.-P.; Hicks, R.; Lever, A.M.L. HIV-1 Gag–RNA Interaction Occurs at a Perinuclear/Centrosomal Site; Analysis by Confocal Microscopy and FRET. Traffic 2005, 6, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Carlson, L.-A.; Bai, Y.; Keane, S.C.; Doudna, J.A.; Hurley, J.H. Reconstitution of Selective HIV-1 RNA Packaging in Vitro by Membrane-Bound Gag Assemblies. eLife 2016, 5, e14663. [Google Scholar] [CrossRef]
- Barajas, B.C.; Tanaka, M.; Robinson, B.A.; Phuong, D.J.; Chutiraka, K.; Reed, J.C.; Lingappa, J.R. Identifying the Assembly Intermediate in Which Gag First Associates with Unspliced HIV-1 RNA Suggests a Novel Model for HIV-1 RNA Packaging. PLoS Pathog. 2018, 14, e1006977. [Google Scholar] [CrossRef]
- Kenyon, J.C.; Prestwood, L.J.; Lever, A.M.L. A Novel Combined RNA-Protein Interaction Analysis Distinguishes HIV-1 Gag Protein Binding Sites from Structural Change in the Viral RNA Leader. Sci. Rep. 2015, 5, 14369. [Google Scholar] [CrossRef]
- Vasa, S.M.; Guex, N.; Wilkinson, K.A.; Weeks, K.M.; Giddings, M.C. ShapeFinder: A Software System for High-Throughput Quantitative Analysis of Nucleic Acid Reactivity Information Resolved by Capillary Electrophoresis. RNA 2008, 14, 1979–1990. [Google Scholar] [CrossRef]
- Aboul-ela, F.; Karn, J.; Varani, G. The Structure of the Human Immunodeficiency Virus Type-1 TAR RNA Reveals Principles of RNA Recognition by Tat Protein. J. Mol. Biol. 1995, 253, 313–332. [Google Scholar] [CrossRef]
- Guo, F.; Saadatmand, J.; Niu, M.; Kleiman, L. Roles of Gag and NCp7 in Facilitating TRNA(Lys)(3) Annealing to Viral RNA in Human Immunodeficiency Virus Type 1. J. Virol. 2009, 83, 8099–8107. [Google Scholar] [CrossRef]
- Saadatmand, J.; Niu, M.; Kleiman, L.; Guo, F. The Contribution of the Primer Activation Signal to Differences between Gag- and NCp7-Facilitated TRNA(Lys3) Annealing in HIV-1. Virology 2009, 391, 334–341. [Google Scholar] [CrossRef]
- Levin, J.G.; Mitra, M.; Mascarenhas, A.; Musier-Forsyth, K. Role of HIV-1 Nucleocapsid Protein in HIV-1 Reverse Transcription. RNA Biol. 2010, 7, 754–774. [Google Scholar] [CrossRef]
- Greatorex, J.; Gallego, J.; Varani, G.; Lever, A. Structure and Stability of Wild-Type and Mutant RNA Internal Loops from the SL-1 Domain of the HIV-1 Packaging Signal. J. Mol. Biol. 2002, 322, 543–557. [Google Scholar] [CrossRef]
- Ennifar, E.; Yusupov, M.; Walter, P.; Marquet, R.; Ehresmann, B.; Ehresmann, C.; Dumas, P. The Crystal Structure of the Dimerization Initiation Site of Genomic HIV-1 RNA Reveals an Extended Duplex with Two Adenine Bulges. Structure 1999, 7, 1439–1449. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Gorelick, R.J.; Vasa, S.M.; Guex, N.; Rein, A.; Mathews, D.H.; Giddings, M.C.; Weeks, K.M. High-Throughput SHAPE Analysis Reveals Structures in HIV-1 Genomic RNA Strongly Conserved across Distinct Biological States. PLoS Biol. 2008, 6, e96. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.C.; Lever, A.M.L. Human Immunodeficiency Virus Type 1 Gag Polyprotein Modulates Its Own Translation. J. Virol. 2006, 80, 10478–10486. [Google Scholar] [CrossRef]
- Tuffy, K.M.; Maldonado, R.J.K.; Chang, J.; Rosenfeld, P.; Cochrane, A.; Parent, L.J. HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites. Viruses 2020, 12, 1281. [Google Scholar] [CrossRef] [PubMed]
- Andersen, E.S.; Contera, S.A.; Knudsen, B.; Damgaard, C.K.; Besenbacher, F.; Kjems, J. Role of the Trans-Activation Response Element in Dimerization of HIV-1 RNA. J. Biol. Chem. 2004, 279, 22243–22249. [Google Scholar] [CrossRef] [PubMed]
- Das, A.T.; Vrolijk, M.M.; Harwig, A.; Berkhout, B. Opening of the TAR Hairpin in the HIV-1 Genome Causes Aberrant RNA Dimerization and Packaging. Retrovirology 2012, 9, 59. [Google Scholar] [CrossRef]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global Changes in the RNA Binding Specificity of HIV-1 Gag Regulate Virion Genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef]
- Damgaard, C.K.; Dyhr-Mikkelsen, H.; Kjems, J. Mapping the RNA Binding Sites for Human Immunodeficiency Virus Type-1 Gag and NC Proteins within the Complete HIV-1 and -2 Untranslated Leader Regions. Nucleic Acids Res. 1998, 26, 3667–3676. [Google Scholar] [CrossRef]
- Guzman, R.N.D.; Wu, Z.R.; Stalling, C.C.; Pappalardo, L.; Borer, P.N.; Summers, M.F. Structure of the HIV-1 Nucleocapsid Protein Bound to the SL3 Ψ-RNA Recognition Element. Science 1998, 279, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Wahab, E.W.; Smyth, R.P.; Mailler, E.; Bernacchi, S.; Vivet-Boudou, V.; Hijnen, M.; Jossinet, F.; Mak, J.; Paillart, J.-C.; Marquet, R. Specific Recognition of the HIV-1 Genomic RNA by the Gag Precursor. Nat. Commun. 2014, 5, 4304. [Google Scholar] [CrossRef] [PubMed]
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. Structure of the HIV-1 RNA Packaging Signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Kharytonchyk, S.; Waller, A.; Mbaekwe, U.; Basappa, S.; Kuo, N.; Frank, H.M.; Quasney, C.; Kidane, A.; Swanson, C.; et al. Identification of the Initial Nucleocapsid Recognition Element in the HIV-1 RNA Packaging Signal. Proc. Natl. Acad. Sci. USA 2020, 117, 17737–17746. [Google Scholar] [CrossRef]
- Bell, N.M.; Kenyon, J.C.; Balasubramanian, S.; Lever, A.M.L. Comparative Structural Effects of HIV-1 Gag and Nucleocapsid Proteins in Binding to and Unwinding of the Viral RNA Packaging Signal. Biochemistry 2012, 51, 3162–3169. [Google Scholar] [CrossRef]
- Bernacchi, S.; El-Wahab, E.W.A.; Dubois, N.; Hijnen, M.; Smyth, R.P.; Mak, J.; Marquet, R.; Paillart, J.-C. HIV-1 Pr55Gag Binds Genomic and Spliced RNAs with Different Affinity and Stoichiometry. RNA Biol. 2017, 14, 90–103. [Google Scholar] [CrossRef]
- Smyth, R.P.; Despons, L.; Huili, G.; Bernacchi, S.; Hijnen, M.; Mak, J.; Jossinet, F.; Weixi, L.; Paillart, J.-C.; von Kleist, M.; et al. Mutational Interference Mapping Experiment (MIME) for Studying RNA Structure and Function. Nat. Methods 2015, 12, 866–872. [Google Scholar] [CrossRef]
- Grohman, J.K.; Gorelick, R.J.; Kottegoda, S.; Allbritton, N.L.; Rein, A.; Weeks, K.M. An Immature Retroviral RNA Genome Resembles a Kinetically Trapped Intermediate State. J. Virol. 2014, 88, 6061–6068. [Google Scholar] [CrossRef]
- Ohishi, M.; Nakano, T.; Sakuragi, S.; Shioda, T.; Sano, K.; Sakuragi, J. The Relationship between HIV-1 Genome RNA Dimerization, Virion Maturation and Infectivity. Nucleic Acids Res. 2011, 39, 3404–3417. [Google Scholar] [CrossRef]
- Shehu-Xhilaga, M.; Crowe, S.M.; Mak, J. Maintenance of the Gag/Gag-Pol Ratio Is Important for Human Immunodeficiency Virus Type 1 RNA Dimerization and Viral Infectivity. J. Virol. 2001, 75, 1834–1841. [Google Scholar] [CrossRef]
- Song, R.; Kafaie, J.; Yang, L.; Laughrea, M. HIV-1 Viral RNA Is Selected in the Form of Monomers That Dimerize in a Three-Step Protease-Dependent Process; the DIS of Stem–Loop 1 Initiates Viral RNA Dimerization. J. Mol. Biol. 2007, 371, 1084–1098. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Souza, A.R.; Jayaraman, D.; Long, Z.; Zeng, J.; Prestwood, L.J.; Chan, C.; Kappei, D.; Lever, A.M.L.; Kenyon, J.C. HIV-1 Packaging Visualised by In-Gel SHAPE. Viruses 2021, 13, 2389. https://doi.org/10.3390/v13122389
D’Souza AR, Jayaraman D, Long Z, Zeng J, Prestwood LJ, Chan C, Kappei D, Lever AML, Kenyon JC. HIV-1 Packaging Visualised by In-Gel SHAPE. Viruses. 2021; 13(12):2389. https://doi.org/10.3390/v13122389
Chicago/Turabian StyleD’Souza, Aaron R., Dhivya Jayaraman, Ziqi Long, Jingwei Zeng, Liam J. Prestwood, Charlene Chan, Dennis Kappei, Andrew M. L. Lever, and Julia C. Kenyon. 2021. "HIV-1 Packaging Visualised by In-Gel SHAPE" Viruses 13, no. 12: 2389. https://doi.org/10.3390/v13122389
APA StyleD’Souza, A. R., Jayaraman, D., Long, Z., Zeng, J., Prestwood, L. J., Chan, C., Kappei, D., Lever, A. M. L., & Kenyon, J. C. (2021). HIV-1 Packaging Visualised by In-Gel SHAPE. Viruses, 13(12), 2389. https://doi.org/10.3390/v13122389





