Phylogenetic Analysis of South African Bovine Leukaemia Virus (BLV) Isolates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Blood Samples and Extraction of Total Bovine Genomic DNA
2.2. Amplification of BLV env Gene by Nested Polymerase Chain Reaction (nPCR) and Sanger Sequencing of Full-Length env Gene
2.3. Amplification of BLV gag Gene by nPCR and Sanger Sequencing of Full-Length gag Gene
2.4. Phylogenetic Analysis of BLV Full-Length and Partial env and Full-Length gag Nucleotide Sequences
2.5. Pairwise Comparison of BLV Env and Gag Nucleotide and Amino Acid Sequence
2.6. Intragenotype and Intergenotype Evolutionary Distances of BLV Env Nucleotide and Amino Acid Sequences
2.7. Alignment of BLV Env and Gag Amino Acid Sequences
3. Results
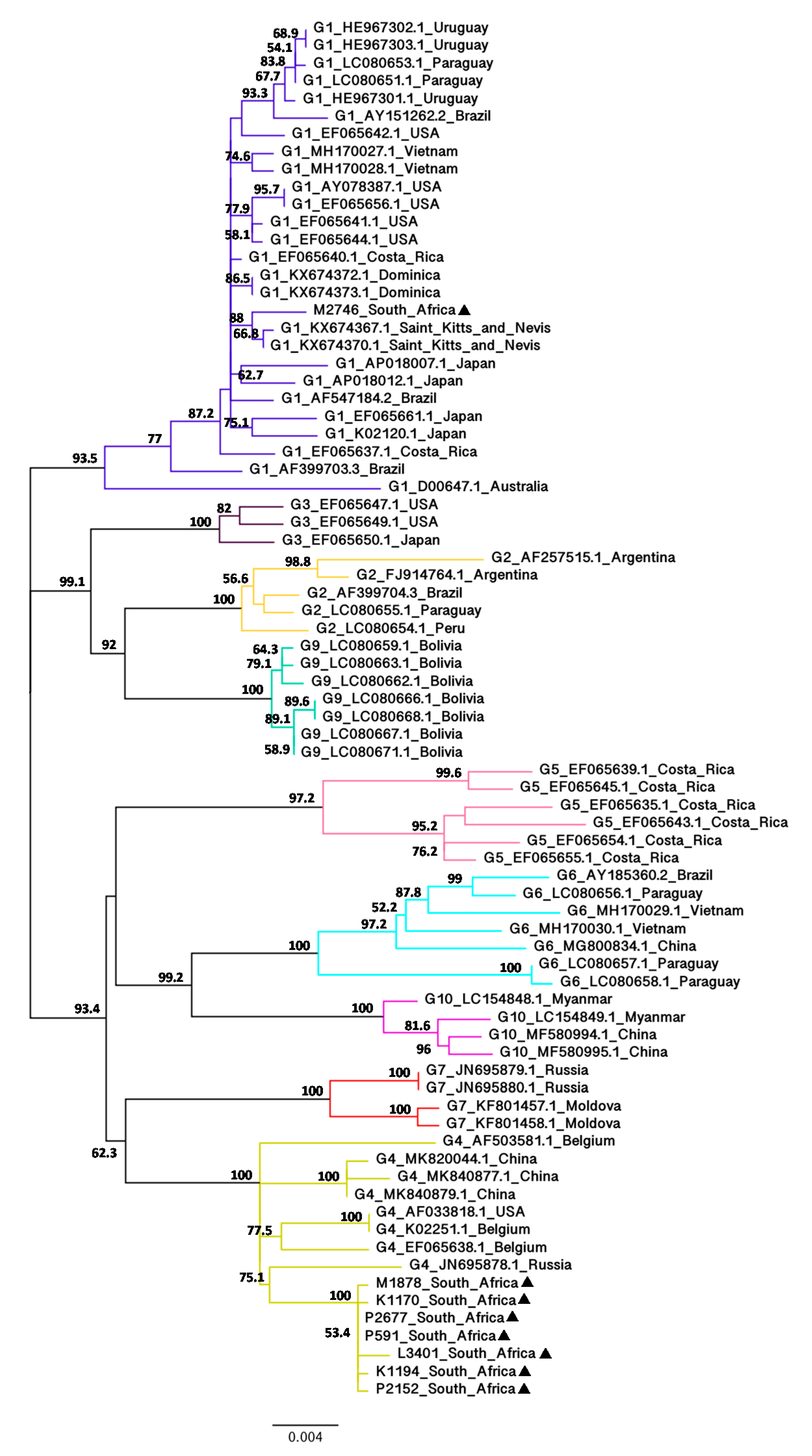
3.1. Phylogenetic Analysis of BLV Full-Length env Nucleotide Sequences
3.2. Phylogenetic Analysis of BLV Partial env Nucleotide Sequences (444 bp)
3.3. Phylogenetic Analysis of BLV Full-Length gag Nucleotide Sequences
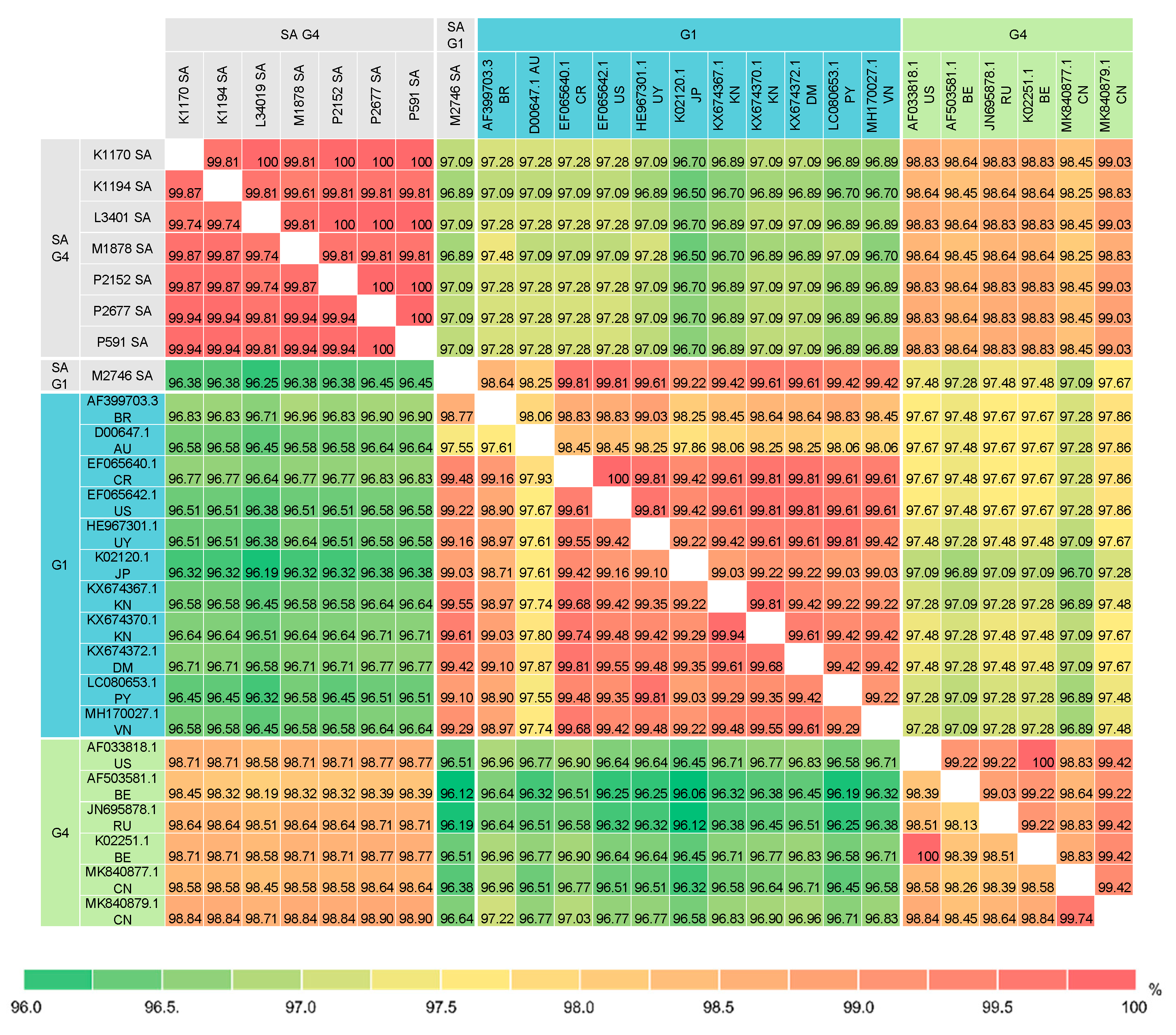
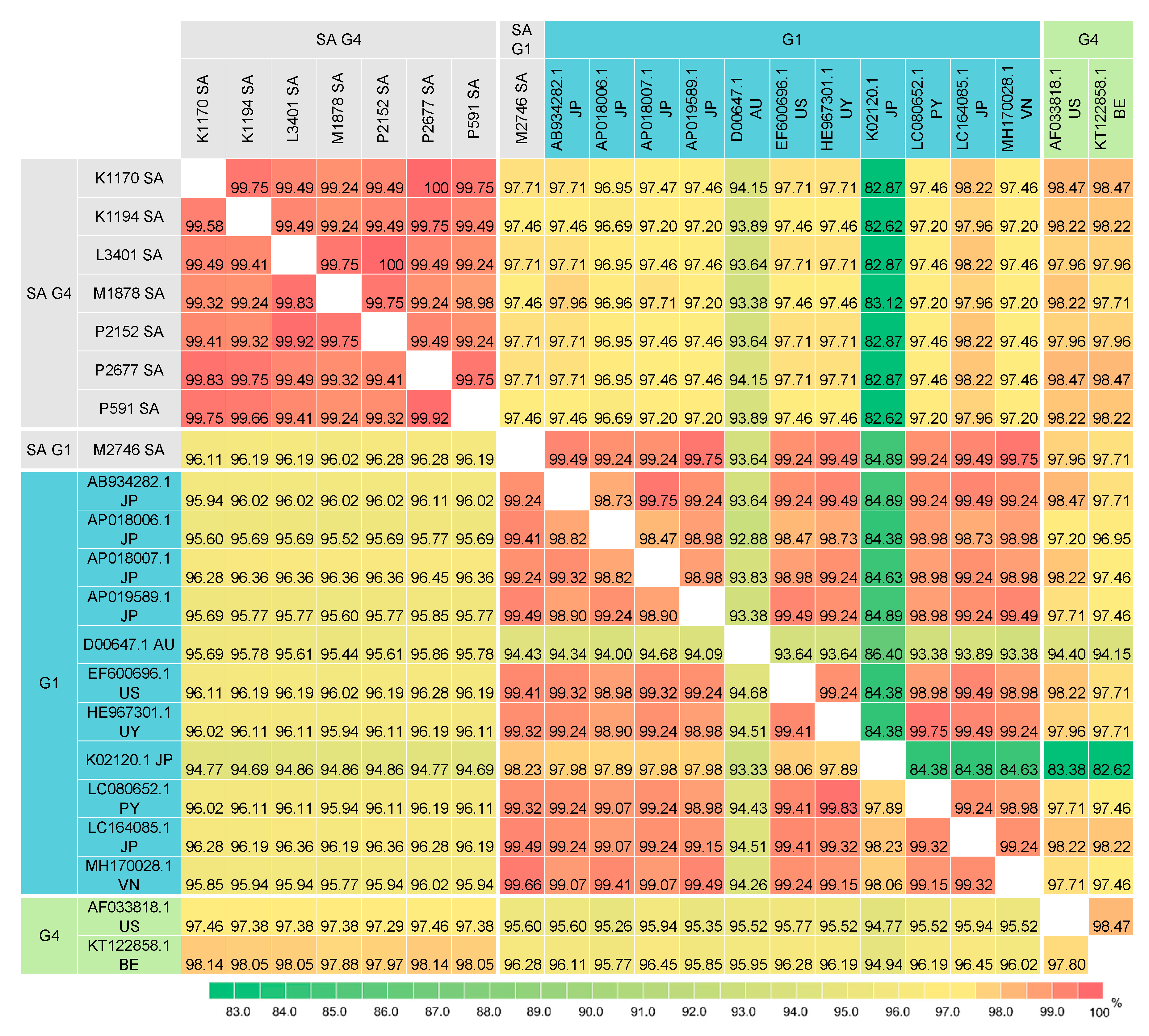
3.4. Pairwise Comparison of the BLV Full-Length env Sequences
3.5. Pairwise Comparison of BLV Partial Env Sequences
3.6. Pairwise Comparison of BLV Gag Sequences
3.7. Genetic Variabilities of BLV Env Sequences
3.8. Alignment of BLV Env Amino Acid Sequences
3.9. Alignment of BLV Gag Amino Acid Sequences
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Onuma, M.; Honma, T.; Mikami, T.; Ichijo, S.; Konishi, T. Studies on the sporadic and enzootic forms of bovine leukosis. J. Comp. Pathol. 1979, 89, 159–167. [Google Scholar] [CrossRef]
- Theilen, G.H.; Dungworth, D.L. Bovine lymphosarcoma in California. 3. Calf form. Am. J. Vet. Res. 1965, 26, 696–709. [Google Scholar] [PubMed]
- Van Der Maaten, M.J.; Miller, J.M.; Boothe, A.D. Replicating type-C virus particles in monolayer cell cultures of tissues from cattle with lymphosarcoma. J. Natl. Cancer Inst. 1974, 52, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Burny, A.; Bruck, C.; Chantrenne, H.; Cleuter, Y.; Dekegel, D.; Ghysdael, J.; Kettmann, R.; Leclercq, M.; Leunen, J.; Mammerickx, M. Bovine leukemia virus: Molecular biology and epidemiology. In Viral Oncology; Klein, G., Ed.; Raven Press: New York, NY, USA, 1980. [Google Scholar]
- DiGiacomo, R.F.; Darlington, R.L.; Evermann, J.F. Natural transmission of bovine leukemia virus in dairy calves by dehorning. Can. J. Comp. Med. Rev. Can. De Med. Comp. 1985, 49, 340–342. [Google Scholar]
- Hopkins, S.G.; DiGiacomo, R.F. Natural Transmission of Bovine Leukemia Virus in Dairy and Beef Cattle. Vet. Clin. N. Am. Food Anim. Pract. 1997, 13, 107–128. [Google Scholar] [CrossRef]
- Lucas, M.H.; Roberts, D.H.; Wibberley, G. Ear tattooing as a method of spread of bovine leukosis virus infection. Br. Vet. J. 1985, 141, 647–649. [Google Scholar] [CrossRef]
- Kohara, J.; Konnai, S.; Onuma, M. Experimental transmission of Bovine leukemia virus in cattle via rectal palpation. Jpn. J. Vet. Sci. 2006, 54, 25–30. [Google Scholar]
- Divers, T.J.; Bartholomew, R.C.; Galligan, D.; Littel, C. Evidence for transmission of bovine leukemia virus by rectal palpation in a commercial dairy herd. Prev. Vet. Med. 1995, 23, 133–141. [Google Scholar] [CrossRef]
- Schwartz, I.; Levy, D. Pathobiology of bovine leukemia virus. Vet. Res. 1994, 25, 521–536. [Google Scholar]
- Yoon, S.S.; Bae, Y.C.; Lee, K.H.; Han, B.; Han, H.R. Characteristics of Bovine Lymphoma Caused by Bovine Leukemia Virus Infection in Holstein-Friesian Dairy Cattle in Korea. Asian-Australas J. Ani. Sci. 2005, 18, 728–733. [Google Scholar] [CrossRef]
- Burny, A.; Bruck, C.; Cleuter, Y.; Couez, D.; Deschamps, J.; Gregoire, D.; Ghysdael, J.; Kettmann, R.; Mammerickx, M.; Marbaix, G.; et al. Bovine leukaemia virus and enzootic bovine leukosis. Onderstepoort J. Vet. Res. 1985, 52, 133–144. [Google Scholar]
- Zarkik, S.; Decroly, E.; Wattiez, R.; Seidah Nabil, G.; Burny, A.; Ruysschaert, J.-M. Comparative processing of bovine leukemia virus envelope glycoprotein gp72 by subtilisin/kexin-like mammalian convertases. Febs Lett. 1997, 406, 205–210. [Google Scholar] [CrossRef] [Green Version]
- Johnston, E.R.; Albritton, L.M.; Radke, K. Envelope Proteins Containing Single Amino Acid Substitutions Support a Structural Model of the Receptor-Binding Domain of Bovine Leukemia Virus Surface Protein. J. Virol. 2002, 76, 10861–10872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatot, J.S.; Callebaut, I.; Van Lint, C.; Demonte, D.; Kerkhofs, P.; Portetelle, D.; Burny, A.; Willems, L.; Kettmann, R. Bovine Leukemia Virus SU Protein Interacts with Zinc, and Mutations within Two Interacting Regions Differently Affect Viral Fusion and Infectivity In Vivo. J. Virol. 2002, 76, 7956–7967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruck, C.; Portetelle, D.; Burny, A.; Zavada, J. Topographical analysis by monoclonal antibodies of BLV-gp51 epitopes involved in viral functions. Virology 1982, 122, 353–362. [Google Scholar] [CrossRef]
- Bruck, C.; Mathot, S.; Portetelle, D.; Berte, C.; Franssen, J.-D.; Herion, P.; Burny, A. Monoclonal antibodies define eight independent antigenic regions on the bovine leukemia virus (BLV) envelope glycoprotein gp51. Virology 1982, 122, 342–352. [Google Scholar] [CrossRef]
- Bruck, C.; Portetelle, D.; Mammerickx, M.; Mathot, S.; Burny, A. Epitopes of bovine leukemia virus glycoprotein gp51 recognized by sera of infected cattle and sheep. Leuk. Res. 1984, 8, 315–321. [Google Scholar] [CrossRef]
- Bruck, C.; Rensonnet, N.; Portetelle, D.; Cleuter, Y.; Mammerickx, M.; Burny, A.; Mamoun, R.; Guillemain, B.; Van Der Maaten, M.J.; Ghysdael, J. Biologically active epitopes of bovine leukemia virus glycoprotein GP51: Their dependence on protein glycosylation and genetic variability. Virology 1984, 136, 20–31. [Google Scholar] [CrossRef]
- Ban, J.; Czene, S.; Altaner, C.; Callebaut, I.; Krchnak, V.; Merza, M.; Burny, A.; Kettmann, R.; Portetelle, D. Mapping of sequential epitopes recognized by monoclonal antibodies on the bovine leukaemia virus external glycoproteins expressed in Escherichia coli by means of antipeptide antibodies. J. Gen. Virol. 1992, 73 Pt 9, 2457–2461. [Google Scholar] [CrossRef]
- Callebaut, I.; Burny, A.; Krchnak, V.; Gras-Masse, H.; Wathelet, B.; Portetelle, D. Use of synthetic peptides to map sequential epitopes recognized by monoclonal antibodies on the bovine leukemia virus external glycoprotein. Virology 1991, 185, 48–55. [Google Scholar] [CrossRef]
- Bai, L.; Takeshima, S.N.; Isogai, E.; Kohara, J.; Aida, Y. Novel CD8(+) cytotoxic T cell epitopes in bovine leukemia virus with cattle. Vaccine 2015, 33, 7194–7202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Brogniez, A.; Mast, J.; Willems, L. Determinants of the bovine leukemia virus envelope glycoproteins involved in infectivity, replication and pathogenesis. Viruses 2016, 8, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callebaut, I.; Voneche, V.; Mager, A.; Fumiere, O.; Krchnak, V.; Merza, M.; Zavada, J.; Mammerickx, M.; Burny, A.; Portetelle, D. Mapping of B-neutralizing and T-helper cell epitopes on the bovine leukemia virus external glycoprotein gp51. J. Virol. 1993, 67, 5321–5327. [Google Scholar] [CrossRef] [Green Version]
- Dube, S.; Abbott, L.; Dube, D.K.; Dolcini, G.; Gutierrez, S.; Ceriani, C.; Juliarena, M.; Ferrer, J.; Perzova, R.; Poiesz, B.J. The complete genomic sequence of an in vivo low replicating BLV strain. Virol. J. 2009, 6, 120. [Google Scholar] [CrossRef] [Green Version]
- Moratorio, G.; Fischer, S.; Bianchi, S.; Tome, L.; Rama, G.; Obal, G.; Carrion, F.; Pritsch, O.; Cristina, J. A detailed molecular analysis of complete bovine leukemia virus genomes isolated from B-cell lymphosarcomas. Vet. Res. 2013, 44, 19. [Google Scholar] [CrossRef] [Green Version]
- Voneche, V.; Portetelle, D.; Kettmann, R.; Willems, L.; Limbach, K.; Paoletti, E.; Ruysschaert, J.M.; Burny, A.; Brasseur, R. Fusogenic segments of bovine leukemia virus and simian immunodeficiency virus are interchangeable and mediate fusion by means of oblique insertion in the lipid bilayer of their target cells. Proc. Natl. Acad. Sci. USA 1992, 89, 3810–3814. [Google Scholar] [CrossRef] [Green Version]
- Willems, L.; Kettmann, R.; Dequiedt, F.; Portetelle, D.; Voneche, V.; Cornil, I.; Kerkhofs, P.; Burny, A.; Mammerickx, M. In vivo infection of sheep by bovine leukemia virus mutants. J. Virol. 1993, 67, 4078–4085. [Google Scholar] [CrossRef] [Green Version]
- Willems, L.; Gatot, J.S.; Mammerickx, M.; Portetelle, D.; Burny, A.; Kerkhofs, P.; Kettmann, R. The YXXL signalling motifs of the bovine leukemia virus transmembrane protein are required for in vivo infection and maintenance of high viral loads. J. Virol. 1995, 69, 4137–4141. [Google Scholar] [CrossRef] [Green Version]
- Reichert, M.; Winnicka, A.; Willems, L.; Kettmann, R.; Cantor, G.H. Role of the proline-rich motif of bovine leukemia virus transmembrane protein gp30 in viral load and pathogenicity in sheep. J. Virol. 2001, 75, 8082–8089. [Google Scholar] [CrossRef] [Green Version]
- Inabe, K.; Nishizawa, M.; Tajima, S.; Ikuta, K.; Aida, Y. The YXXL sequences of a transmembrane protein of bovine leukemia virus are required for viral entry and incorporation of viral envelope protein into virions. J. Virol. 1999, 73, 1293–1301. [Google Scholar] [CrossRef] [Green Version]
- Portetelle, D.; Limbach, K.; Burny, A.; Mammerickx, M.; Desmettre, P.; Riviere, M.; Zavada, J.; Paoletti, E. Recombinant vaccinia virus expression of the bovine leukaemia virus envelope gene and protection of immunized sheep against infection. Vaccine 1991, 9, 194–200. [Google Scholar] [CrossRef]
- Kumar, S.; Andrew, M.E.; Boyle, D.B.; Brandon, R.B.; Lavin, M.F.; Daniel, R.C. Expression of bovine leukaemia virus envelope gene by recombinant vaccinia viruses. Virus Res. 1990, 17, 131–142. [Google Scholar] [CrossRef]
- Gillet, N.; Florins, A.; Boxus, M.; Burteau, C.; Nigro, A.; Vandermeers, F.; Balon, H.; Bouzar, A.B.; Defoiche, J.; Burny, A.; et al. Mechanisms of leukemogenesis induced by bovine leukemia virus: Prospects for novel anti-retroviral therapies in human. Retrovirology 2007, 4, 18. [Google Scholar] [CrossRef] [Green Version]
- Rice, N.R.; Stephens, R.M.; Burny, A.; Gilden, R.V. The gag and pol genes of bovine leukemia virus: Nucleotide sequence and analysis. Virology 1985, 142, 357–377. [Google Scholar] [CrossRef]
- Llames, L.; Goyache, J.; Domenech, A.; Montaña, A.V.; Suarez, G.; Gomez-Lucia, E. Cellular distribution of bovine leukemia virus proteins gp51SU, Pr72env, and Pr66gag-pro in persistently infected cells. Virus Res. 2001, 79, 47–57. [Google Scholar] [CrossRef]
- Yoshinaka, Y.; Katoh, I.; Copeland, T.D.; Smythers, G.W.; Oroszlan, S. Bovine leukemia virus protease: Purification, chemical analysis, and in vitro processing of gag precursor polyproteins. J. Virol. 1986, 57, 826–832. [Google Scholar] [CrossRef] [Green Version]
- Juliarena, M.A.; Gutierrez, S.E.; Ceriani, C. Determination of proviral load in bovine leukemia virus-infected cattle with and without lymphocytosis. Am. J. Vet. Res. 2007, 68, 1220–1225. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. WAHIS Interface Animal Health Situation: Disease Distribution Maps. Available online: https://www.oie.int/wahis_2/public/wahid.php/Diseaseinformation/Diseasedistributionmap?disease_type_hidden=&disease_id_hidden=&selected_disease_name_hidden=&disease_type=0&disease_id_terrestrial=35&species_t=0&disease_id_aquatic=-999&species_a=0&sta_method=semesterly&selected_start_year=2016&selected_report_period=1&selected_start_month=12&date_submit=OK (accessed on 4 January 2020).
- European Commission. Bovine and Swine Diseases Situation 2017; European Commission: Brussels, Belgium, 2017; pp. 1–35. [Google Scholar]
- Rodriguez, S.M.; Golemba, M.D.; Campos, R.H.; Trono, K.; Jones, L.R. Bovine leukemia virus can be classified into seven genotypes: Evidence for the existence of two novel clades. J. Gen. Virol. 2009, 90, 2788–2797. [Google Scholar] [CrossRef]
- Balic, D.; Lojkic, I.; Periskic, M.; Bedekovic, T.; Jungic, A.; Lemo, N.; Roic, B.; Cac, Z.; Barbic, L.; Madic, J. Identification of a new genotype of bovine leukemia virus. Arch. Virol. 2012, 157, 1281–1290. [Google Scholar] [CrossRef]
- Rola-Luszczak, M.; Pluta, A.; Olech, M.; Donnik, I.; Petropavlovskiy, M.; Gerilovych, A.; Vinogradova, I.; Choudhury, B.; Kuzmak, J. The molecular characterization of bovine leukaemia virus isolates from Eastern Europe and Siberia and its impact on phylogeny. PLoS ONE 2013, 8, e58705. [Google Scholar] [CrossRef]
- Polat, M.; Takeshima, S.N.; Hosomichi, K.; Kim, J.; Miyasaka, T.; Yamada, K.; Arainga, M.; Murakami, T.; Matsumoto, Y.; de la Barra Diaz, V.; et al. A new genotype of bovine leukemia virus in South America identified by NGS-based whole genome sequencing and molecular evolutionary genetic analysis. Retrovirology 2016, 13, 4. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Kim, E.J.; Ratthanophart, J.; Vitoonpong, R.; Kim, B.H.; Cho, I.S.; Song, J.Y.; Lee, K.K.; Shin, Y.K. Molecular epidemiological and serological studies of bovine leukemia virus (BLV) infection in Thailand cattle. Infect Genet Evol. 2016, 41, 245–254. [Google Scholar] [CrossRef]
- Polat, M.; Moe, H.H.; Shimogiri, T.; Moe, K.K.; Takeshima, S.N.; Aida, Y. The molecular epidemiological study of bovine leukemia virus infection in Myanmar cattle. Arch. Virol. 2017, 162, 425–437. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Baloch, A.R.; Pan, Y.; Xu, F.; Tian, L.; Zeng, Q. Molecular epidemiology and characterization of bovine leukemia virus in domestic yaks (Bos grunniens on the Qinghai-Tibet Plateau, China. Arch. Virol. 2018, 163, 659–670. [Google Scholar] [CrossRef]
- Yu, C.; Wang, X.; Zhou, Y.; Wang, Y.; Zhang, X.; Zheng, Y. Genotyping bovine leukemia virus in dairy cattle of Heilongjiang, northeastern China. BMC Vet. Res. 2019, 15. [Google Scholar] [CrossRef]
- Murakami, K.; Kobayashi, S.; Konishi, M.; Kameyama, K.; Yamamoto, T.; Tsutsui, T. The recent prevalence of bovine leukemia virus (BLV) infection among Japanese cattle. Vet. Microbiol. 2011, 148, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Trono, K.G.; Perez-Filgueira, D.M.; Duffy, S.; Borca, M.V.; Carrillo, C. Seroprevalence of bovine leukemia virus in dairy cattle in Argentina: Comparison of sensitivity and specificity of different detection methods. Vet. Microbiol. 2001, 83, 235–248. [Google Scholar] [CrossRef]
- Monti, G.; Schrijver, R.; Beier, D. Genetic diversity and spread of Bovine leukaemia virus isolates in Argentine dairy cattle. Arch. Virol. 2005, 150, 443–458. [Google Scholar] [CrossRef]
- Kobayashi, S.; Tsutsui, T.; Yamamoto, T.; Hayama, Y.; Kameyama, K.; Konishi, M.; Murakami, K. Risk factors associated with within-herd transmission of bovine leukemia virus on dairy farms in Japan. BMC Vet. Res. 2010, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Sandev, N.; Ilieva, D.; Sizov, I.; Rusenova, N.; Iliev, E. Prevalence of Enzootic Bovine Leukosis in the Republic of Bulgaria in 1997–2004. Vet. Arh. 2006, 76, 263–268. [Google Scholar]
- Bauermann, F.V.; Ridpath, J.F.; Dargatz, D.A. Bovine leukemia virus seroprevalence among cattle presented for slaughter in the United States. J. Vet. Diagn. Investig. Off. Publ. Am. Assoc. Vet. Lab. Diagn. Inc. 2017, 29, 704–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaDronka, R.M.; Ainsworth, S.; Wilkins, M.J.; Norby, B.; Byrem, T.M.; Bartlett, P.C. Prevalence of Bovine Leukemia Virus Antibodies in US Dairy Cattle. Vet. Med. Int. 2018, 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Polat, M.; Takeshima, S.-n.; Aida, Y. Epidemiology and genetic diversity of bovine leukemia virus. Virol. J. 2017, 14. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. Int. J. Org. Evol. 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.S.; Simulundu, E.; Mwiinga, D.; Samui, K.L.; Mweene, A.S.; Kajihara, M.; Mangani, A.; Mwenda, R.; Ndebe, J.; Konnai, S.; et al. Clinical and subclinical bovine leukemia virus infection in a dairy cattle herd in Zambia. Arch. Virol. 2017, 162, 1051–1056. [Google Scholar] [CrossRef] [PubMed]
- Phiri, M.M.; Kaimoyo, E.; Changula, K.; Silwamba, I.; Chambaro, H.M.; Kapila, P.; Kajihara, M.; Simuunza, M.; Muma, J.B.; Pandey, G.S.; et al. Molecular detection and characterization of genotype 1 bovine leukemia virus from beef cattle in the traditional sector in Zambia. Arch. Virol. 2019, 164, 2531–2536. [Google Scholar] [CrossRef]
- Lee, E.; Kim, E.J.; Joung, H.K.; Kim, B.H.; Song, J.Y.; Cho, I.S.; Lee, K.K.; Shin, Y.K. Sequencing and phylogenetic analysis of the gp51 gene from Korean bovine leukemia virus isolates. Virol. J. 2015, 12, 64. [Google Scholar] [CrossRef] [Green Version]
- Felmer, R.; Munoz, G.; Zuniga, J.; Recabal, M. Molecular analysis of a 444 bp fragment of the bovine leukaemia virus gp51 env gene reveals a high frequency of non-silent point mutations and suggests the presence of two subgroups of BLV in Chile. Vet. Microbiol. 2005, 108, 39–47. [Google Scholar] [CrossRef]
- Ababneh, M.M.; Al-Rukibat, R.K.; Hananeh, W.M.; Nasar, A.T.; Al-Zghoul, M.B. Detection and molecular characterization of bovine leukemia viruses from Jordan. Arch. Virol. 2012, 157, 2343–2348. [Google Scholar] [CrossRef]
- Asfaw, Y.; Tsuduku, S.; Konishi, M.; Murakami, K.; Tsuboi, T.; Wu, D.; Sentsui, H. Distribution and superinfection of bovine leukemia virus genotypes in Japan. Arch. Virol. 2005, 150, 493–505. [Google Scholar] [CrossRef]
- Bicka, L.; Kuzmak, J.; Rola, M.; Beier, D. Detection of genetic diversity among bovine leukemia virus population by single-strand conformational polymorphism analysis. Bull. Vet. Inst. Pulawy 2002, 46, 205–212. [Google Scholar] [CrossRef]
- Fechner, H.; Blankenstein, P.; Looman, A.C.; Elwert, J.; Geue, L.; Albrecht, C.; Kurg, A.; Beier, D.; Marquardt, O.; Ebner, D. Provirus variants of the bovine leukemia virus and their relation to the serological status of naturally infected cattle. Virology 1997, 237, 261–269. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Buehring, G.C. Natural genetic variations in bovine leukemia virus envelope gene: Possible effects of selection and escape. Virology 2007, 366, 150–165. [Google Scholar] [CrossRef] [Green Version]
- Pluta, A.; Albritton, L.M.; Rola-Luszczak, M.; Kuzmak, J. Computational analysis of envelope glycoproteins from diverse geographical isolates of bovine leukemia virus identifies highly conserved peptide motifs. Retrovirology 2018, 15, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pluta, A.; Rola-Luszczak, M.; Kubis, P.; Balov, S.; Moskalik, R.; Choudhury, B.; Kuzmak, J. Molecular characterization of bovine leukemia virus from Moldovan dairy cattle. Arch. Virol. 2017, 162, 1563–1576. [Google Scholar] [CrossRef] [Green Version]
- Portetelle, D.; Couez, D.; Bruck, C.; Kettmann, R.; Mammerickx, M.; Van der Maaten, M.; Brasseur, R.; Burny, A. Antigenic variants of bovine leukemia virus (BLV) are defined by amino acid substitutions in the NH2 part of the envelope glycoprotein gp51. Virology 1989, 169, 27–33. [Google Scholar] [CrossRef]
- Portetelle, D.; Dandoy, C.; Burny, A.; Zavada, J.; Siakkou, H.; Gras-Masse, H.; Drobecq, H.; Tartar, A. Synthetic peptides approach to identification of epitopes on bovine leukemia virus envelope glycoprotein gp51. Virology 1989, 169, 34–41. [Google Scholar] [CrossRef]
- Mamoun, R.Z.; Morisson, M.; Rebeyrotte, N.; Busetta, B.; Couez, D.; Kettmann, R.; Hospital, M.; Guillemain, B. Sequence variability of bovine leukemia virus env gene and its relevance to the structure and antigenicity of the glycoproteins. J. Virol. 1990, 64, 4180–4188. [Google Scholar] [CrossRef] [Green Version]
- Moratorio, G.; Obal, G.; Dubra, A.; Correa, A.; Bianchi, S.; Buschiazzo, A.; Cristina, J.; Pritsch, O. Phylogenetic analysis of bovine leukemia viruses isolated in South America reveals diversification in seven distinct genotypes. Arch. Virol. 2010, 155, 481–489. [Google Scholar] [CrossRef]
- Camargos, M.F.; Pereda, A.; Stancek, D.; Rocha, M.A.; dos Reis, J.K.; Greiser-Wilke, I.; Leite, R.C. Molecular characterization of the env gene from Brazilian field isolates of Bovine leukemia virus. Virus Genes 2007, 34, 343–350. [Google Scholar] [CrossRef]
- Auerbach, M.R.; Shu, C.; Kaplan, A.; Singh, I.R. Functional characterization of a portion of the Moloney murine leukemia virus gag gene by genetic footprinting. Proc. Natl. Acad. Sci. USA 2003, 100, 11678. [Google Scholar] [CrossRef] [Green Version]
- Morcock, D.; Kane, B.; Casas-Finet, J. Fluorescence and nucleic acid binding properties of the human T-cell leukemia virus-type 1 nucleocapsid protein. Biochim. Et. Biophys. Acta 2000, 1481, 381–394. [Google Scholar] [CrossRef]
- Fu, W.; Hu, W.-S. Functional Replacement of Nucleocapsid Flanking Regions by Heterologous Counterparts with Divergent Primary Sequences: Effects of Chimeric Nucleocapsid on the Retroviral Replication Cycle. J. Virol. 2003, 77, 754–761. [Google Scholar] [CrossRef] [Green Version]
- Gorelick, R.J.; Henderson, L.E.; Hanser, J.P.; Rein, A. Point Mutants of Moloney Murine Leukemia Virus that Fail to Package Viral RNA: Evidence for Specific RNA Recognition by a Zinc FingerLike Protein Sequence. Proc. Natl. Acad. Sci. USA 1988, 85, 8420–8424. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, M.D.; Fiore, D.; Panganiban, A.T. Distinct functions and requirements for the Cys-His boxes of the human immunodeficiency virus type 1 nucleocapsid protein during RNA encapsidation and replication. J. Virol. 1997, 71, 9295–9305. [Google Scholar] [CrossRef] [Green Version]
- Bowzard, J.B.; Bennett, R.P.; Krishna, N.K.; Ernst, S.M.; Rein, A.; Wills, J.W. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 1998, 72, 9034–9044. [Google Scholar] [CrossRef] [Green Version]
- Poon, D.T.; Wu, J.; Aldovini, A. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 1996, 70, 6607–6616. [Google Scholar] [CrossRef] [Green Version]
- Schmalzbauer, E.; Strack, B.; Dannull, J.; Guehmann, S.; Moelling, K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J. Virol. 1996, 70, 771–777. [Google Scholar] [CrossRef] [Green Version]
- Dannull, J.; Surovoy, A.; Jung, G.; Moelling, K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. Embo J. 1994, 13, 1525–1533. [Google Scholar] [CrossRef]
- Jewell, N.A.; Mansky, L.M. In the beginning: Genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J. Gen. Virol. 2000, 81, 1889–1899. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Norris, K.M.; Mansky, L.M. Involvement of the matrix and nucleocapsid domains of the bovine leukemia virus Gag polyprotein precursor in viral RNA packaging. J. Virol. 2003, 77, 9431–9438. [Google Scholar] [CrossRef] [Green Version]



| gp51 | gp30 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Epitopes and Motifs | Leader Sequence | G Epitope | H Epitope | G Epitope | ND2 | D Epitope | PXXP Motif | |||||||||||
| Amino acid position | 29 | 48 | 56 | 59 | 73 | 74 | 82 | 121 | 132 | 134 | 144 | 153 | 254 | 476 | 479 | 480 | 504 | |
| References | d | a,d | a | a,b | a,d | a,d | a,c | a | a,d | a,d | a,d | d | d | d | ||||
| SA G1 | M2746 | R | A | S | I | A | K | S | R | Q | D | I | Q | S | E | L | T | V |
| G1 | AF399703.3 BR | Q | A | S | I | A | K | F | R | Q | N | I | H | S | E | S | T | V |
| AP018021.1 JP | R | A | S | I | A | K | S | R | Q | D | I | H | S | E | L | T | V | |
| EF065641.1 US | R | A | S | I | A | K | S | R | Q | D | I | H | S | E | L | T | V | |
| EF065646.1 JP | Q | A | S | I | A | K | S | R | Q | D | I | H | S | E | L | T | V | |
| HE967301.1 UY | R | A | S | I | A | K | S | R | Q | N | I | H | S | E | L | T | V | |
| KX674639.1 KN | R | A | S | I | A | K | S | R | Q | D | I | H | S | E | L | T | V | |
| LC440653.1 ZM | − | − | − | − | − | − | − | R | Q | D | I | H | − | − | − | − | − | |
| LC075558.1 PY | − | − | − | − | − | − | − | R | Q | D | I | H | − | − | − | − | − | |
| G2 | AF257515.1 AR | Q | A | S | I | A | K | F | R | Q | D | I | H | L | E | L | A | T |
| LC080654.1 PE | Q | V | S | I | A | K | F | R | Q | D | I | H | L | E | F | A | T | |
| LC080655.1 PY | Q | A | S | I | A | K | F | R | Q | D | I | H | L | E | F | A | T | |
| G3 | EF065647.1 US | Q | A | S | I | A | K | F | R | Q | D | I | H | L | E | F | A | T |
| EF065650.1 JP | Q | A | S | I | A | K | F | R | Q | D | I | H | L | E | F | A | T | |
| JQ686091.1 RU | Q | T | S | I | P | R | F | H | Q | D | I | H | L | E | F | P | T | |
| SA G4 | K1170 | Q | T | F | I | P | R | F | H | R | D | T | H | L | D | F | P | T |
| K1194 | Q | T | F | L | P | R | F | H | R | D | T | H | L | D | F | P | T | |
| L3401 | Q | T | F | I | A | R | F | H | R | D | T | H | L | D | F | P | T | |
| M1878 | Q | T | F | I | P | R | F | H | R | N | T | H | L | D | F | P | T | |
| P2152 | Q | T | F | I | P | R | F | H | R | D | T | H | L | D | F | P | T | |
| P2677 | Q | T | F | I | P | R | F | H | R | D | T | H | L | D | F | P | T | |
| P591 | Q | T | F | I | P | R | F | H | R | D | T | H | L | D | F | P | T | |
| G4 | AF033818.1 US | Q | T | S | I | P | R | F | H | Q | D | I | H | L | E | F | P | T |
| AF067081.1 PL | − | − | − | − | − | − | − | H | Q | D | I | H | − | − | − | − | − | |
| AY515279.1 CL | Q | T | S | I | P | R | F | H | Q | D | I | H | L | E | F | P | S | |
| JN695878.1 RU | Q | T | S | I | P | R | F | H | Q | D | I | H | L | E | F | P | T | |
| KF801460.2 MD | Q | T | S | I | P | R | F | H | Q | D | I | H | L | E | F | P | T | |
| LC193462.1 ZM | − | − | − | − | − | − | − | H | R | D | T | H | − | − | − | − | − | |
| M35238.1 FR | Q | T | F | I | P | R | F | H | Q | D | T | H | L | E | F | P | T | |
| M35240.1 BE | Q | T | S | I | P | R | F | H | Q | D | I | H | L | E | F | P | T | |
| MK820044.1 CN | Q | T | S | I | P | R | F | H | Q | D | I | H | L | E | F | P | T | |
| G5 | AF399702.3 BR | Q | T | S | I | A | R | F | R | R | D | I | H | L | − | − | − | − |
| EF065635.1 CR | R | T | S | I | A | R | F | R | R | D | I | H | L | E | F | T | T | |
| EF065636.1 CR | R | T | S | I | A | R | F | R | R | D | I | H | L | E | L | T | V | |
| G6 | AY185360.2 BR | Q | T | S | I | A | R | F | R | Q | D | T | H | L | E | F | T | T |
| LC075576.1 BO | Q | T | S | I | A | R | F | R | Q | D | T | H | L | E | F | T | T | |
| LC080656.1 PY | Q | T | S | I | A | R | F | R | Q | D | T | H | L | E | F | T | T | |
| MF580991.1 CN | Q | T | S | I | A | R | F | R | Q | D | T | Y | L | E | F | T | T | |
| KU233530.1 TH | − | − | − | − | − | − | − | R | Q | D | T | H | − | − | − | − | − | |
| G7 | KF801457.1.MD | Q | I | S | I | A | R | F | R | Q | D | I | H | L | E | F | T | A |
| JN695879.1 RU | Q | T | S | I | A | R | F | R | Q | D | I | H | L | E | F | T | A | |
| S83530.1 IT | − | − | − | − | − | − | − | R | Q | D | I | H | − | − | − | − | − | |
| G8 | GU724606.1 HR | − | − | − | − | − | − | − | R | Q | D | I | H | − | − | − | − | − |
| JF713455.1 RU | − | − | − | − | − | − | − | R | Q | D | I | H | − | − | − | − | − | |
| LT970927.1 IT | − | − | − | − | − | − | − | R | Q | D | I | H | − | − | − | − | − | |
| G9 | LC080659.1 BO | Q | A | S | I | A | K | L | R | Q | D | I | H | L | E | F | A | T |
| LC080664.1 BO | Q | A | S | I | A | K | F | R | Q | D | I | H | L | E | F | A | T | |
| G10 | KU233527.1 TH | − | − | − | − | − | − | − | H | Q | D | T | H | − | − | − | − | − |
| LC154848.1 MM | Q | T | S | I | A | R | F | R | Q | D | T | H | L | E | F | T | T | |
| MF580994.1 CN | Q | T | S | I | A | R | F | H | Q | D | T | H | L | E | F | T | T | |
| G11 | KU764746.1 CN | − | − | − | − | − | − | − | R | Q | D | I | H | − | − | − | − | − |
| p15 MA | p24 CA | p12 NC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino Acid Position | 29 | 63 | 69 | 88 | 107 | 108 | 278 | 318 | 341 | 343 | 365 | |
| References | a | a | a | a | ||||||||
| SA G1 | M2746 | N | T | K | G | A | V | I | M | K | P | A |
| G1 | AP018021.1 JP | N | T | R | G | A | V | I | M | K | P | A |
| D00647.1 AU | N | A | K | E | A | V | I | T | K | P | T | |
| AB934282.1 JP | N | T | R | G | A | I | I | M | K | P | A | |
| LC005615.1 JP | N | T | R | G | A | V | I | M | K | P | A | |
| LC080651.1 PY | N | T | R | G | A | V | I | M | K | P | A | |
| MH170027.1 VN | N | T | R | G | A | V | I | M | K | P | A | |
| MH170028.1 VN | N | T | R | G | A | V | I | M | K | P | A | |
| EF600696.1 US | N | T | R | G | A | V | I | V | K | P | A | |
| HE967301.1 UY | N | T | R | G | A | V | I | M | K | P | A | |
| G2 | FJ914764.1 AR | N | T | R | G | A | V | I | I | K | P | T |
| LC080654.1 PE | N | T | R | G | A | V | I | I | K | P | T | |
| LC080655.1 PY | N | T | R | G | A | V | I | I | K | P | T | |
| SA G4 | K1170 | D | A | K | E | A | V | V | I | K | S | T |
| K1194 | D | A | K | E | V | V | V | I | K | S | T | |
| L3401 | D | A | K | G | S | V | V | I | K | S | T | |
| M1878 | D | A | R | G | S | I | V | I | K | S | T | |
| P2152 | D | A | K | G | S | V | V | I | K | S | T | |
| P2677 | D | A | K | E | A | V | V | I | K | S | T | |
| P591 | D | A | K | E | A | V | V | I | Q | S | T | |
| G4 | KT122858.1 BE | N | A | K | E | A | V | I | I | K | P | T |
| AF033818.1 US | N | A | K | E | A | I | I | V | K | P | T | |
| G6 | MF580991.1 CN | N | V | K | E | V | V | I | T | K | P | T |
| LC080656.1 PY | N | V | K | E | A | V | I | T | K | P | T | |
| MH170030.1 VN | N | V | K | E | A | I | I | T | K | P | T | |
| G9 | LC080659.1 BO | N | T | R | G | A | V | I | I | K | P | T |
| LC080664.1 BO | N | T | R | G | A | V | I | I | K | P | T | |
| G10 | MF580994.1 CN | N | V | K | E | A | D | I | V | K | P | A |
| LC154848.1 MM | N | V | K | E | A | V | I | V | K | P | A | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, A.; Chapman, R.; Douglass, N.; Carulei, O.; van Rensburg, J.; Williamson, A.-L. Phylogenetic Analysis of South African Bovine Leukaemia Virus (BLV) Isolates. Viruses 2020, 12, 898. https://doi.org/10.3390/v12080898
Suzuki A, Chapman R, Douglass N, Carulei O, van Rensburg J, Williamson A-L. Phylogenetic Analysis of South African Bovine Leukaemia Virus (BLV) Isolates. Viruses. 2020; 12(8):898. https://doi.org/10.3390/v12080898
Chicago/Turabian StyleSuzuki, Akiko, Rosamund Chapman, Nicola Douglass, Olivia Carulei, Johan van Rensburg, and Anna-Lise Williamson. 2020. "Phylogenetic Analysis of South African Bovine Leukaemia Virus (BLV) Isolates" Viruses 12, no. 8: 898. https://doi.org/10.3390/v12080898





