Differential Accumulation of Innate- and Adaptive-Immune-Response-Derived Transcripts during Antagonism between Papaya Ringspot Virus and Papaya Mosaic Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus and Plant Materials
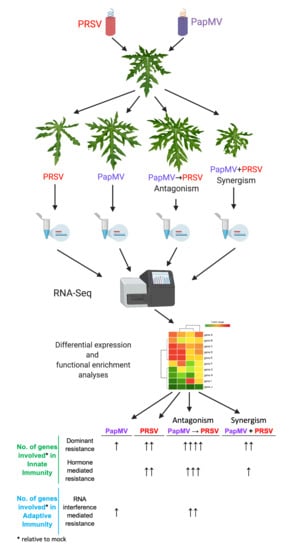
2.2. Experimental Design
2.3. Illumina Sequencing
2.4. De Novo Assembly, Mapping, and Statistical Analysis
2.5. Functional Annotation
2.6. Gene Expression Validation
3. Results
3.1. Symptom Development with Single and Mixed Infections of PapMV and PRSV
3.2. RNA-Seq of Virus-Infected Papaya Plants from Single and Mixed Infections
3.3. Gene Expression Profiling and Functional Annotation Addresses Antagonism Highly Enriched in Immune-Related Response
4. Discussion
4.1. Potyvirus PRSV Triggers Innate Immunity and Potexvirus PapMV Triggers Adaptive Immunity
4.2. Antagonism and Synergism: Similar Expression, Large Differences
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicaise, V. Crop immunity against viruses: Outcomes and future challenges. Front. Plant Sci. 2014, 5, 660. [Google Scholar] [CrossRef] [PubMed]
- De Ronde, D.; Butterbach, P.; Kormelink, R. Dominant resistance against plant viruses. Front. Plant Sci. 2014, 5, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soosaar, J.L.M.; Burch-Smith, T.M.; Dinesh-Kumar, S.P. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 2005, 3, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Aranda, M.A. Recessive resistance to plant viruses. Adv. Virus Res. 2009, 75, 119–159. [Google Scholar]
- Schmitt-Keichinger, C. Manipulating Cellular Factors to Combat Viruses: A Case Study from the Plant Eukaryotic Translation Initiation Factors eIF4. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.Y.; Shirasu, K.; Moon, J.S.; Lee, S.-G.; Kwon, S.-Y. The Activated SA and JA Signaling Pathways Have an Influence on flg22-Triggered Oxidative Burst and Callose Deposition. PLoS ONE 2014, 9, e88951. [Google Scholar] [CrossRef] [Green Version]
- Boller, T. Ethylene in Pathogenesis and Disease Resistance. Available online: https://www.taylorfrancis.com/ (accessed on 12 September 2019).
- Kachroo, P.; Yoshioka, K.; Shah, J.; Dooner, H.K.; Klessig, D.F. Resistance to Turnip Crinkle Virus in Arabidopsis Is Regulated by Two Host Genes and Is Salicylic Acid Dependent but NPR1, Ethylene, and Jasmonate Independent. Plant Cell 2000, 12, 677–690. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Choi, G.H.; Nuss, D.L. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. USA 2009, 106, 17927–17932. [Google Scholar] [CrossRef] [Green Version]
- Zvereva, A.S.; Pooggin, M.M. Silencing and Innate Immunity in Plant Defense Against Viral and Non-Viral Pathogens. Viruses 2012, 4, 2578–2597. [Google Scholar] [CrossRef] [Green Version]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicaise, V.; Candresse, T. Plum pox virus capsid protein suppresses plant pathogen-associated molecular pattern (PAMP)-triggered immunity. Mol. Plant Pathol. 2017, 18, 878–886. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, B.C.; Calil, I.P.; Machado, J.P.B.; Santos, A.A.; Fontes, E.P.B. Immune Receptors and Co-receptors in Antiviral Innate Immunity in Plants. Front. Microbiol. 2017, 7, 2137. [Google Scholar] [CrossRef] [Green Version]
- Roth, B.M.; Pruss, G.J.; Vance, V.B. Plant viral suppressors of RNA silencing. Virus Res. 2004, 102, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Bologna, N.G.; Voinnet, O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014, 65, 473–503. [Google Scholar] [CrossRef]
- Kamitani, M.; Nagano, A.J.; Honjo, M.N.; Kudoh, H. RNA-Seq reveals virus-virus and virus-plant interactions in nature. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascia, T.; Gallitelli, D. Synergies and antagonisms in virus interactions. Plant Sci. 2016, 252, 176–192. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J. Plant Virus Metagenomics: Biodiversity and Ecology. Annu. Rev. Genet. 2012, 46, 359–369. [Google Scholar] [CrossRef]
- Wille, M.; Eden, J.-S.; Shi, M.; Klaassen, M.; Hurt, A.C.; Holmes, E.C. Virus-virus interactions and host ecology are associated with RNA virome structure in wild birds. Mol. Ecol. 2018, 27, 5263–5278. [Google Scholar] [CrossRef]
- Alcalá-Briseño, R.I.; Casarrubias-Castillo, K.; López-Ley, D.; Garrett, K.A.; Silva-Rosales, L. Network analysis of the papaya orchard virome from two agroecological regions of Chiapas, Mexico. bioRxiv 2019, 5, 708479. [Google Scholar] [CrossRef] [Green Version]
- Edwards, R.A.; Rohwer, F. Viral metagenomics. Nat. Rev. Microbiol. 2005, 3, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Syller, J. Facilitative and antagonistic interactions between plant viruses in mixed infections. Mol. Plant Pathol. 2012, 13, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Syller, J.; Grupa, A. Antagonistic within-host interactions between plant viruses: Molecular basis and impact on viral and host fitness. Mol. Plant Pathol. 2016, 17, 769–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruss, G.; Ge, X.; Shi, X.M.; Carrington, J.C.; Vance, V.B. Plant viral synergism: The potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 1997, 9, 859–868. [Google Scholar] [CrossRef]
- Rybicki, E.P. A Top Ten list for economically important plant viruses. Arch. Virol. 2015, 160, 17–20. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Scholthof, K.-B.G. Characterization of a Viral Synergism in the Monocot Brachypodium distachyon Reveals Distinctly Altered Host Molecular Processes Associated with Disease. Plant Physiol. 2012, 160, 1432–1452. [Google Scholar] [CrossRef] [Green Version]
- García-Marcos, A.; Pacheco, R.; Martiáñez, J.; González-Jara, P.; Díaz-Ruíz, J.R.; Tenllado, F. Transcriptional Changes and Oxidative Stress Associated with the Synergistic Interaction Between Potato virus X and Potato virus Y and Their Relationship with Symptom Expression. MPMI 2009, 22, 1431–1444. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Zhao, L.; Yan, B.; Zhu, Y.; Ma, H.; Chen, W.; Ruan, S. Comparative Transcriptome Analysis Reveals the Transcriptional Alterations in Growth- and Development-Related Genes in Sweet Potato Plants Infected and Non-Infected by SPFMV, SPV2, and SPVG. Int. J. Mol. Sci. 2019, 20, 1012. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, S.; Suzuki, J.Y.; Ferreira, S.A.; Gonsalves, D. Papaya ringspot virus-P: Characteristics, pathogenicity, sequence variability and control. Mol. Plant Pathol. 2008, 9, 269–280. [Google Scholar] [CrossRef]
- Chávez-Calvillo, G.; Contreras-Paredes, C.A.; Mora-Macias, J.; Noa-Carrazana, J.C.; Serrano-Rubio, A.A.; Dinkova, T.D.; Carrillo-Tripp, M.; Silva-Rosales, L. Antagonism or synergism between Papaya ringspot virus and Papaya mosaic virus in Carica papaya is determined by their order of infection. Virology 2016, 489, 179–191. [Google Scholar] [CrossRef] [Green Version]
- Fabbro, C.D.; Scalabrin, S.; Morgante, M.; Giorgi, F.M. An Extensive Evaluation of Read Trimming Effects on Illumina NGS Data Analysis. PLoS ONE 2013, 8, e85024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-Seq: Reference generation and analysis with Trinity. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Pimentel, H.; Bray, N.L.; Pachter, L. Gene-level differential analysis at transcript-level resolution. Genome Biol. 2018, 19, 53. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [Green Version]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Tabas-Madrid, D.; Nogales-Cadenas, R.; Pascual-Montano, A. GeneCodis3: A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012, 40, W478–W483. [Google Scholar] [CrossRef]
- Vance, V.V. Replication of Potato virus X RNA is altered in coinfections with Potato virus Y. Virology 1991, 182, 486–494. [Google Scholar]
- Noa-Carrazana, J.C.; González-de-León, D.; Ruiz-Castro, B.S.; Piñero, D.; Silva-Rosales, L. Distribution of Papaya ringspot virus and Papaya mosaic virus in Papaya Plants (Carica papaya) in Mexico. Plant Dis. 2006, 90, 1004–1011. [Google Scholar] [CrossRef] [Green Version]
- Hillung, J.; Cuevas, J.M.; Elena, S.F.P. Transcript Profiling of Different Arabidopsis thaliana Ecotypes in Response to Tobacco etch potyvirus Infection. Front. Microbiol. 2012, 3, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Shang, J.; Wang, W.; Du, J.; Li, K.; Wu, X.; Yu, L.; Liu, C.; Khaskheli, M.I.; Yang, W. Comparison of Transcriptome Differences in Soybean Response to Soybean Mosaic Virus under Normal Light and in the Shade. Viruses 2019, 11, 793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anjanappa, R.B.; Mehta, D.; Okoniewski, M.J.; Szabelska-Berȩsewicz, A.; Gruissem, W.; Vanderschuren, H. Molecular insights into Cassava brown streak virus susceptibility and resistance by profiling of the early host response. Mol. Plant Pathol. 2018, 19, 476–489. [Google Scholar] [CrossRef] [Green Version]
- Zuriaga, E.; Romero, C.; Blanca, J.M.; Badenes, M.L. Resistance to Plum Pox Virus (PPV) in apricot (Prunus armeniaca L.) is associated with down-regulation of two MATHd genes. BMC Plant Biol. 2018, 18, 25. [Google Scholar] [CrossRef] [Green Version]
- Bengyella, L.; Waikhom, S.D.; Allie, F.; Rey, C. Virus tolerance and recovery from viral induced-symptoms in plants are associated with transcriptome reprograming. Plant Mol. Biol. 2015, 89, 243–252. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Zamora, A.; Azhar, M.T.; Sacco, M.A.; Lambert, L.H.; Moffett, P. Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control. Plant J. 2009, 58, 940–951. [Google Scholar] [CrossRef]
- Jaubert, M.; Bhattacharjee, S.; Mello, A.F.S.; Perry, K.L.; Moffett, P. ARGONAUTE2 mediates RNA-silencing antiviral defenses against Potato virus X in Arabidopsis. Plant Physiol. 2011, 156, 1556–1564. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-B.; Lee, H.-Y.; Choi, E.-H.; Park, E.; Kim, J.-H.; Moon, K.-B.; Kim, H.-S.; Choi, D. The Coiled-Coil and Leucine-Rich Repeat Domain of the Potyvirus Resistance Protein Pvr4 Has a Distinct Role in Signaling and Pathogen Recognition. Mol. Plant Microbe Interact. 2018, 31, 906–913. [Google Scholar] [CrossRef] [Green Version]
- Boller, T.; He, S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef] [Green Version]
- Kangasjärvi, S.; Neukermans, J.; Li, S.; Aro, E.-M.; Noctor, G. Photosynthesis, photorespiration, and light signalling in defence responses. J. Exp. Bot. 2012, 63, 1619–1636. [Google Scholar] [CrossRef] [Green Version]
- Karpinski, S.; Gabrys, H.; Mateo, A.; Karpinska, B.; Mullineaux, P.M. Light perception in plant disease defence signalling. Curr. Opin. Plant Biol. 2003, 6, 390–396. [Google Scholar] [CrossRef]
- Madroñero, J.; Rodrigues, S.P.; Antunes, T.F.S.; Abreu, P.M.V.; Ventura, J.A.; Fernandes, A.A.R.; Fernandes, P.M.B. Transcriptome analysis provides insights into the delayed sticky disease symptoms in Carica papaya. Plant Cell Rep. 2018, 37, 967–980. [Google Scholar] [CrossRef] [PubMed]
- Rioja, C.; Wees, S.C.V.; Charlton, K.A.; Pieterse, C.M.J.; Lorenzo, O.; García-Sánchez, S. Wide Screening of Phage-Displayed Libraries Identifies Immune Targets in Planta. PLoS ONE 2013, 8, e54654. [Google Scholar] [CrossRef] [PubMed]
- Pré, M.; Atallah, M.; Champion, A.; Vos, M.D.; Pieterse, C.M.J.; Memelink, J. The AP2/ERF Domain Transcription Factor ORA59 Integrates Jasmonic Acid and Ethylene Signals in Plant Defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [Green Version]
- Folimonova, S.Y. Developing an understanding of cross-protection by Citrus tristeza virus. Front. Microbiol. 2013, 4, 76. [Google Scholar] [CrossRef] [Green Version]
- Endres, M.W.; Gregory, B.D.; Gao, Z.; Foreman, A.W.; Mlotshwa, S.; Ge, X.; Pruss, G.J.; Ecker, J.R.; Bowman, L.H.; Vance, V. Two Plant Viral Suppressors of Silencing Require the Ethylene-Inducible Host Transcription Factor RAV2 to Block RNA Silencing. PLoS Pathog. 2010, 6, e1000729. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Qu, F. Cross Protection of Plant Viruses: Recent Developments and Mechanistic Implications. In Current Research Topics in Plant Virology; Wang, A., Zhou, X., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 241–250. [Google Scholar]
- Folimonova, S.Y. Superinfection Exclusion Is an Active Virus-Controlled Function That Requires a Specific Viral Protein. J. Virol. 2012, 86, 5554–5561. [Google Scholar] [CrossRef] [Green Version]
- Untiveros, M.; Fuentes, S.; Salazar, L.F. Synergistic Interaction of Sweet potato chlorotic stunt virus (Crinivirus) with Carla-, Cucumo-, Ipomo-, and Potyviruses Infecting Sweet Potato. Plant Dis. 2007, 91, 669–676. [Google Scholar] [CrossRef] [Green Version]
- Wamaitha, M.J.; Nigam, D.; Maina, S.; Stomeo, F.; Wangai, A.; Njuguna, J.N.; Holton, T.A.; Wanjala, B.W.; Wamalwa, M.; Lucas, T.; et al. Metagenomic analysis of viruses associated with maize lethal necrosis in Kenya. Virol. J. 2018, 15, 90. [Google Scholar] [CrossRef]





| Condition | Innate Immunity | Adaptive Immunity | ||
|---|---|---|---|---|
| Dominant resistance | Recessive resistance | Hormone mediated resistance | RNA interference mediated resistance | |
| PapMV | ● (12) | N/A * | ● (3) | |
| PRSV | ●● (25) | N/A * | ●● (15) | |
| PapMV→PRSV (Antagonism) | ●●●● (76) | N/A * | ●●● (33) | ●● (15) |
| PapMV + PRSV (Synergism) | ●● (16) | N/A * | ● (5) | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas-Mejía, P.; Vega-Arreguín, J.; Chávez-Calvillo, G.; Ibarra-Laclette, E.; Silva-Rosales, L. Differential Accumulation of Innate- and Adaptive-Immune-Response-Derived Transcripts during Antagonism between Papaya Ringspot Virus and Papaya Mosaic Virus. Viruses 2020, 12, 230. https://doi.org/10.3390/v12020230
Vargas-Mejía P, Vega-Arreguín J, Chávez-Calvillo G, Ibarra-Laclette E, Silva-Rosales L. Differential Accumulation of Innate- and Adaptive-Immune-Response-Derived Transcripts during Antagonism between Papaya Ringspot Virus and Papaya Mosaic Virus. Viruses. 2020; 12(2):230. https://doi.org/10.3390/v12020230
Chicago/Turabian StyleVargas-Mejía, Pablo, Julio Vega-Arreguín, Gabriela Chávez-Calvillo, Enrique Ibarra-Laclette, and Laura Silva-Rosales. 2020. "Differential Accumulation of Innate- and Adaptive-Immune-Response-Derived Transcripts during Antagonism between Papaya Ringspot Virus and Papaya Mosaic Virus" Viruses 12, no. 2: 230. https://doi.org/10.3390/v12020230