Structural Insights on Retroviral DNA Integration: Learning from Foamy Viruses
Abstract
:1. Introduction
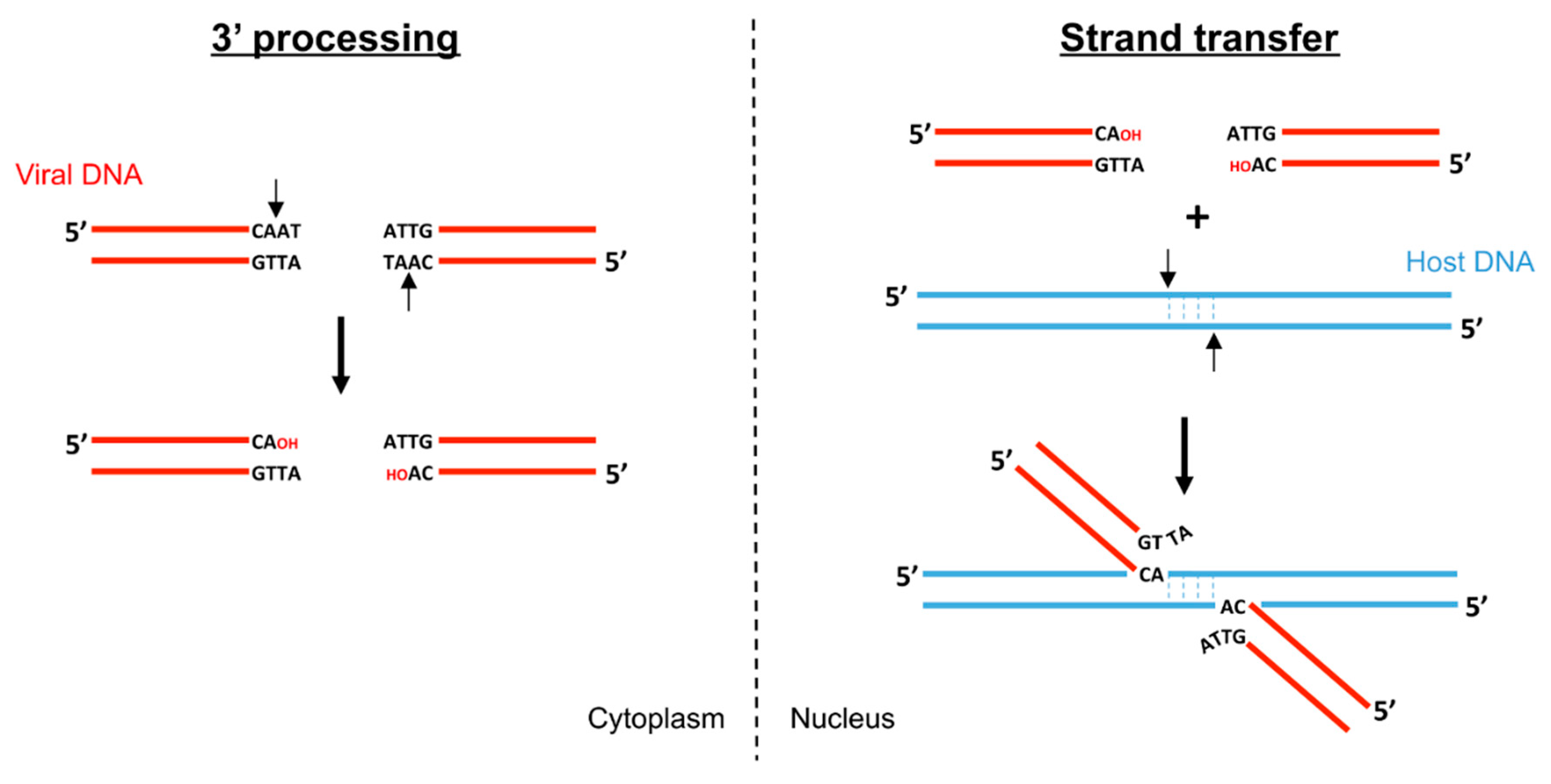
2. Biochemistry of Foamy Virus Integration
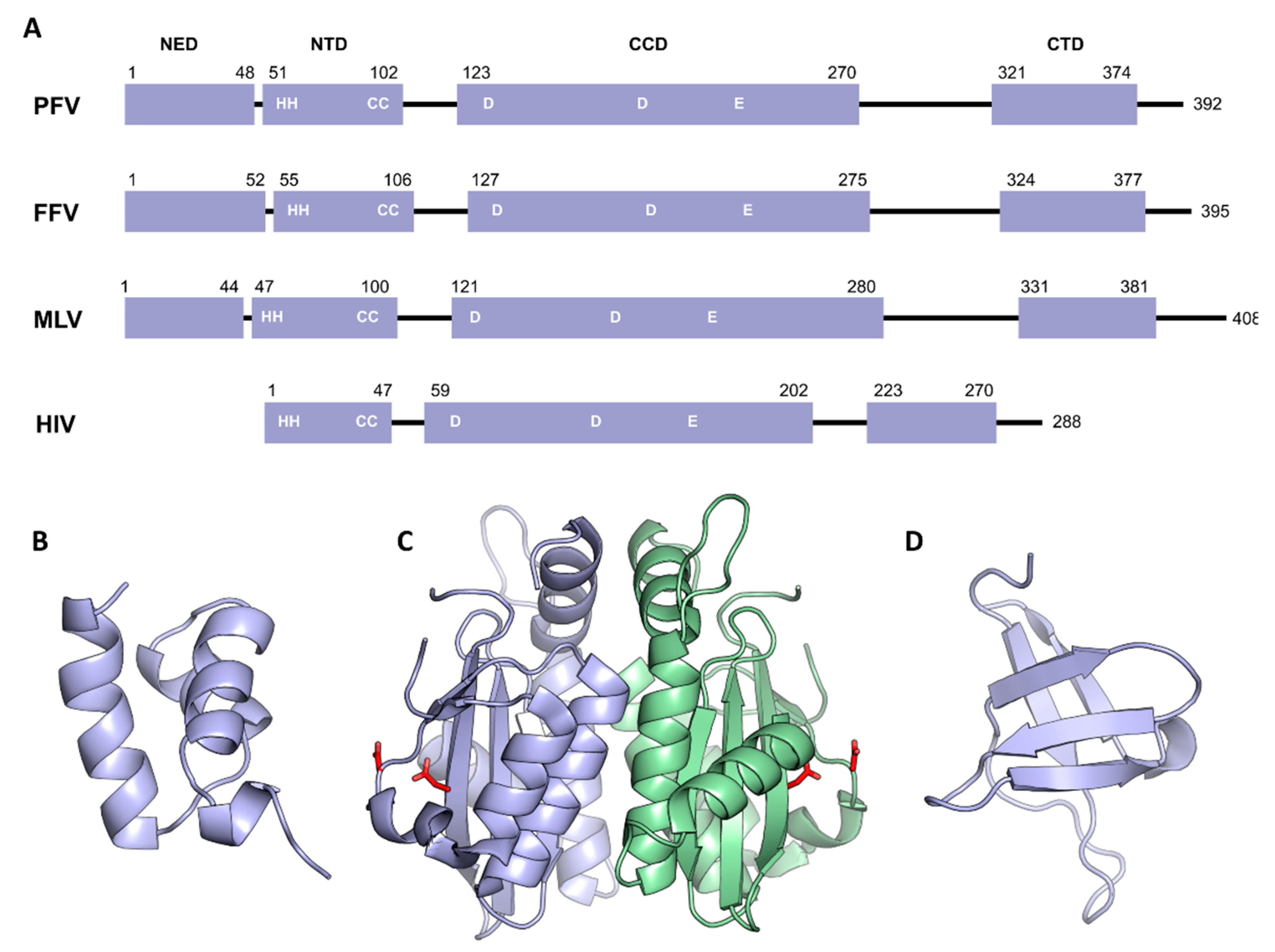
3. Domain Organization of Retroviral Integrase
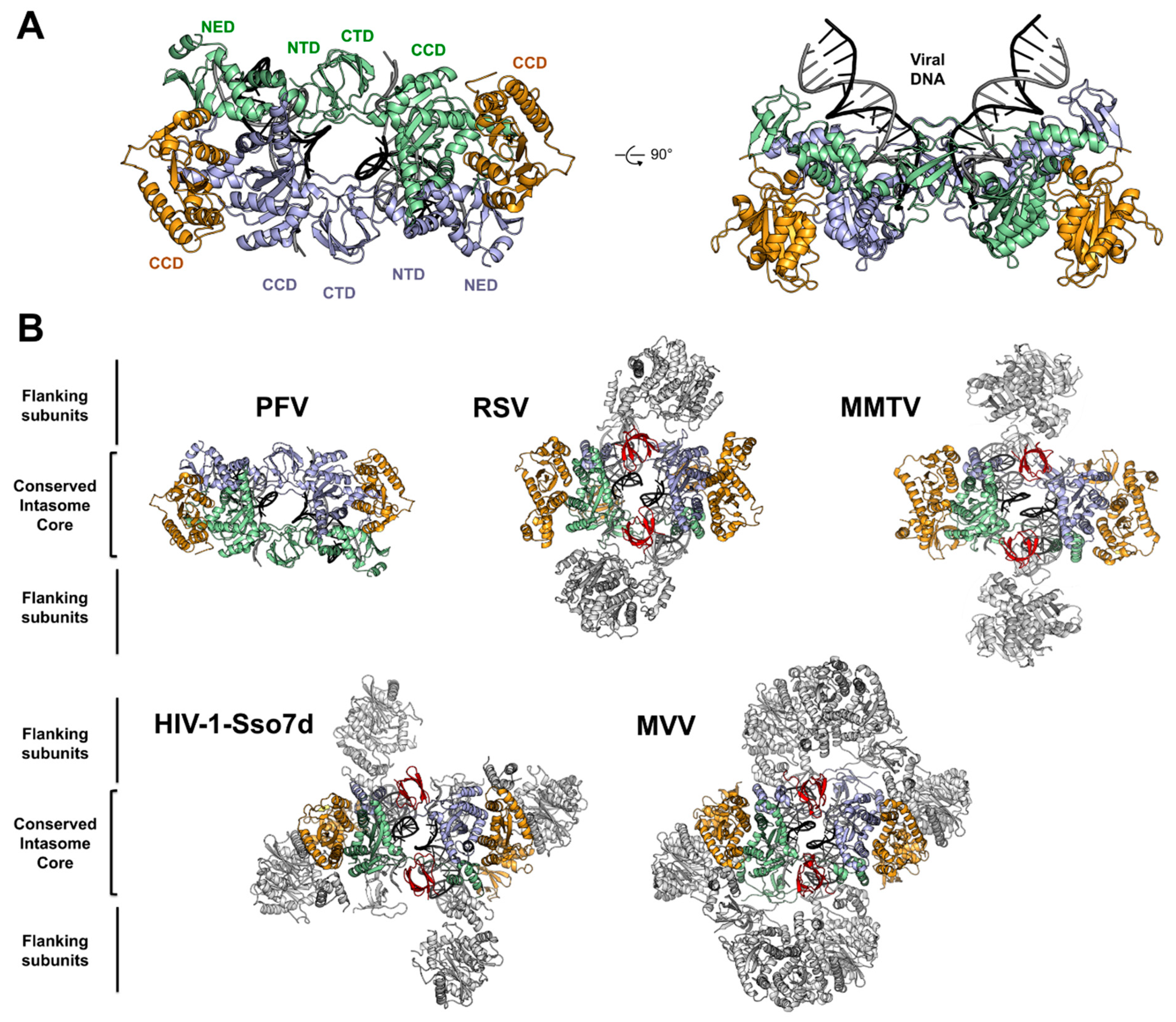
4. Architecture of the PFV Intasome
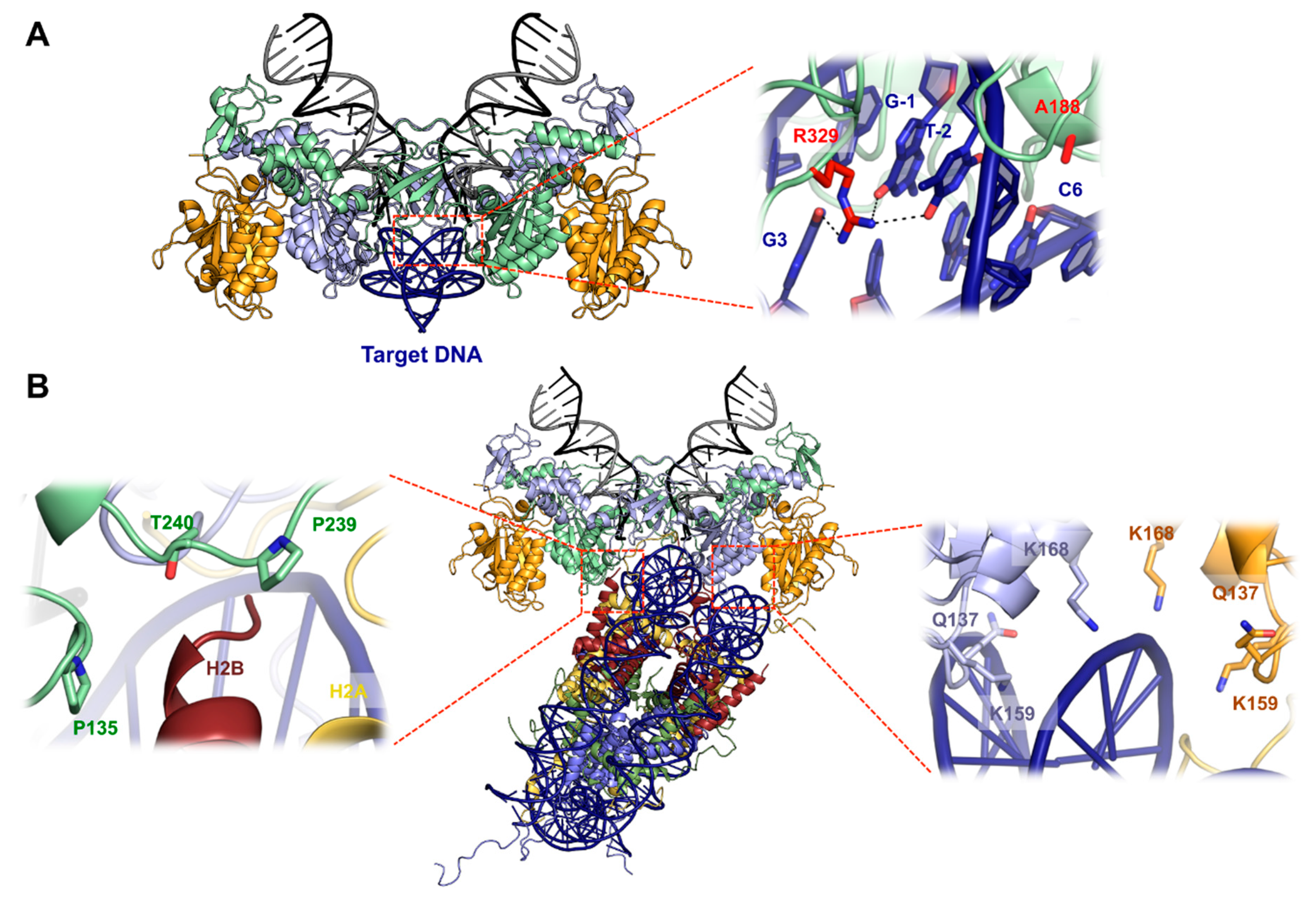
5. Structural Basis for Target DNA Capture
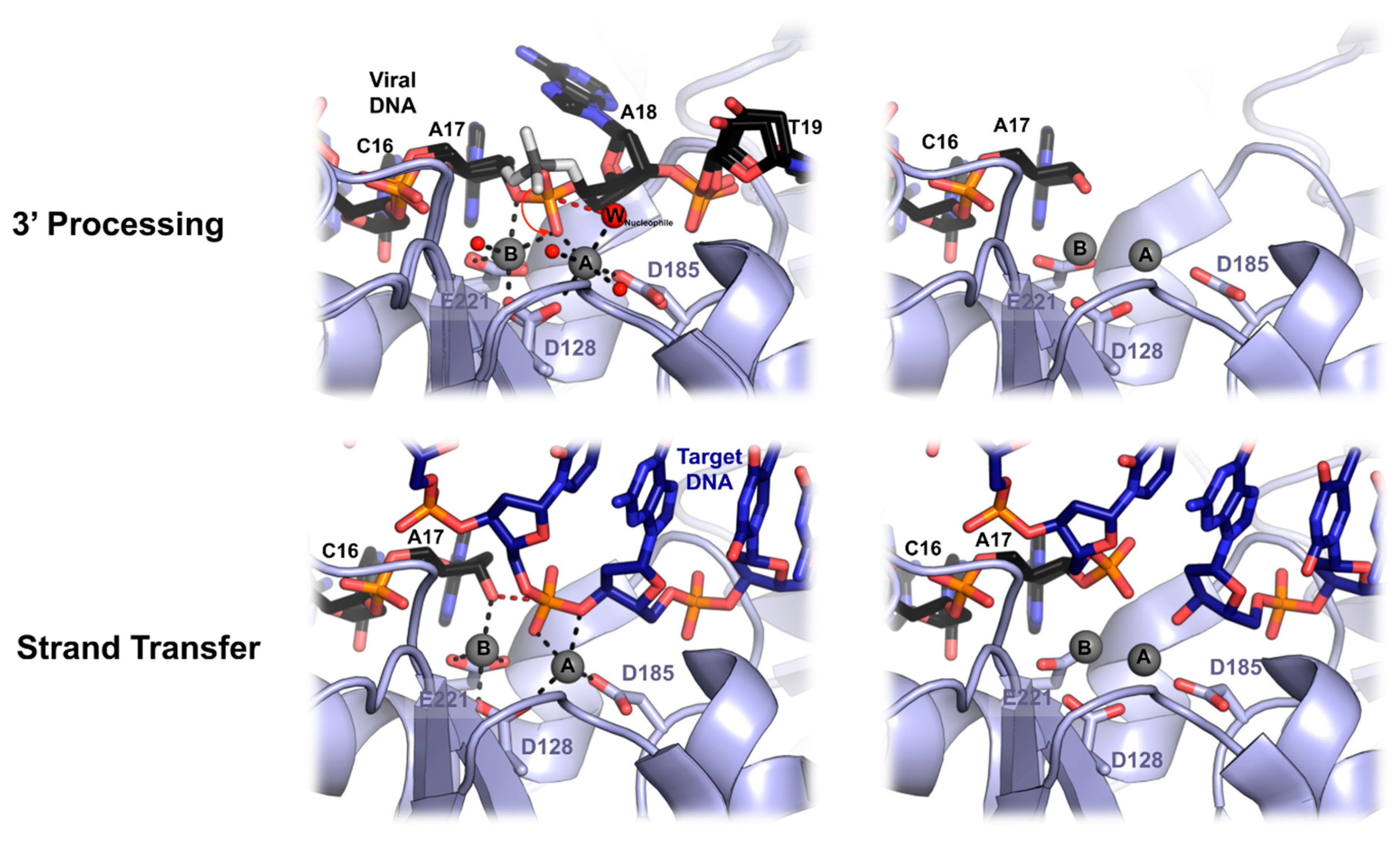
6. Mechanics of PFV Intasome Active Site
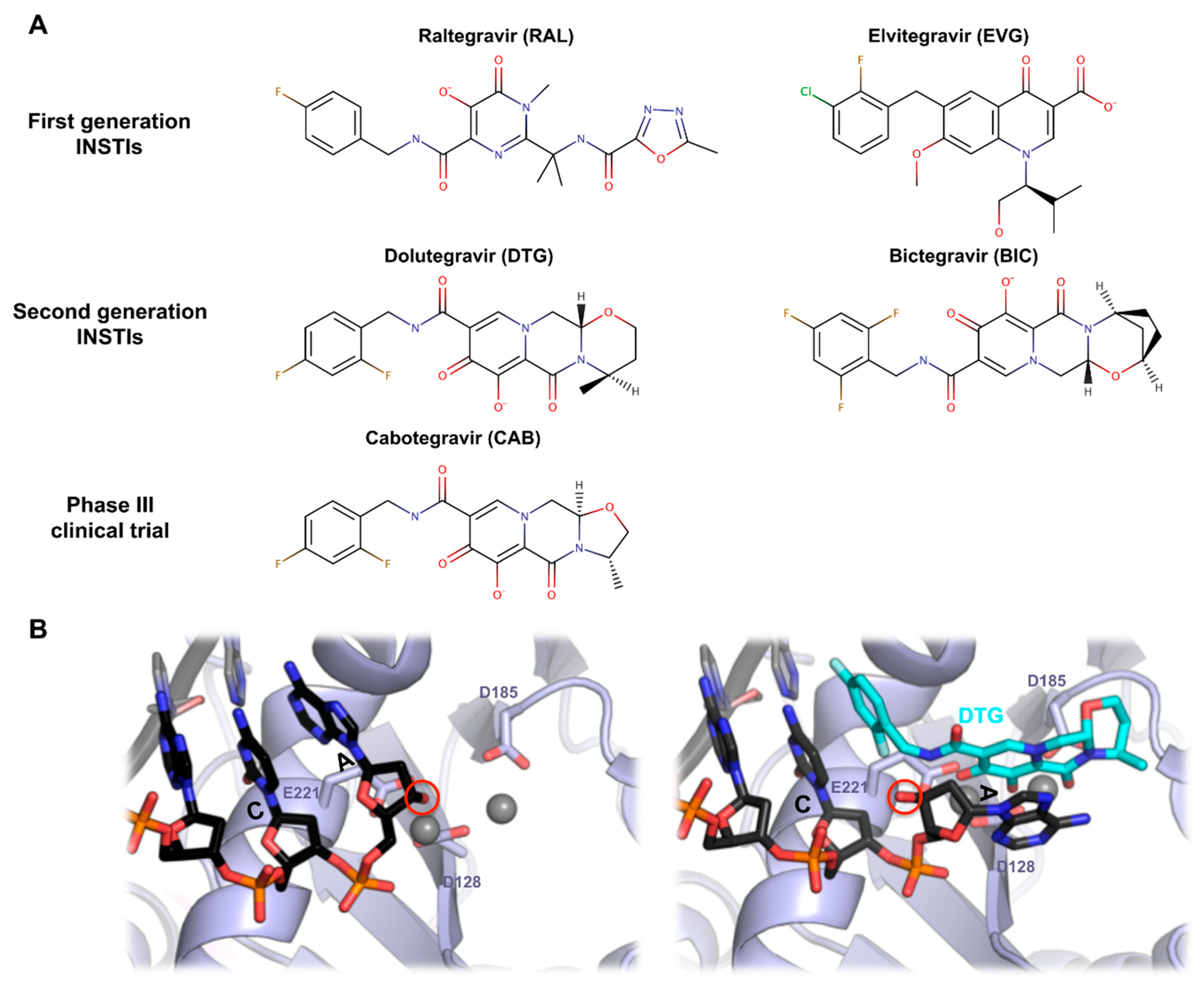
7. PFV Intasome and HIV-1 Strand Transfer Inhibitors
8. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Lesbats, P.; Engelman, A.N.; Cherepanov, P. Retroviral DNA Integration. Chem. Rev. 2016, 116, 12730–12757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dyda, F.; Chandler, M.; Hickman, A.B. The emerging diversity of transpososome architectures. Q. Rev. Biophys. 2012, 45, 493–521. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Mizuuchi, K. Retroviral DNA integration: Structure of an integration intermediate. Cell 1988, 54, 497–504. [Google Scholar] [CrossRef]
- Brown, P.O.; Bowerman, B.; Varmus, H.E.; Bishop, J.M. Retroviral integration: Structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl. Acad. Sci. USA 1989, 86, 2525–2529. [Google Scholar] [CrossRef] [PubMed]
- Roth, M.J.; Schwartzberg, P.L.; Goff, S.P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: Dependence on IN function and terminal DNA sequence. Cell 1989, 58, 47–54. [Google Scholar] [CrossRef]
- Bowerman, B.; Brown, P.O.; Bishop, J.M.; Varmus, H.E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989, 3, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.-Q.; Mizuuchi, K.; Craigie, R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997, 16, 7511–7520. [Google Scholar] [CrossRef]
- Valkov, E.; Gupta, S.S.; Hare, S.; Helander, A.; Roversi, P.; McClure, M.; Cherepanov, P. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009, 37, 243–255. [Google Scholar] [CrossRef]
- Hare, S.; Gupta, S.S.; Valkov, E.; Engelman, A.; Cherepanov, P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 2010, 464, 232–236. [Google Scholar]
- Maertens, G.N.; Hare, S.; Cherepanov, P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 2010, 468, 326–329. [Google Scholar] [Green Version]
- Maskell, D.P.; Renault, L.; Serrao, E.; Lesbats, P.; Matadeen, R.; Hare, S.; Lindemann, D.; Engelman, A.N.; Costa, A.; Cherepanov, P. Structural basis for retroviral integration into nucleosomes. Nature 2015, 523, 366–369. [Google Scholar] [CrossRef] [Green Version]
- Brown, P.O.; Bowerman, B.; Varmus, H.E.; Bishop, J. Correct integration of retroviral DNA in vitro. Cell 1987, 49, 347–356. [Google Scholar] [CrossRef]
- Farnet, C.M.; Haseltine, W.A. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. USA 1990, 87, 4164–4168. [Google Scholar] [CrossRef] [PubMed]
- Skinner, L.M.; Sudol, M.; Harper, A.L.; Katzman, M. Nucleophile selection for the endonuclease activities of human, ovine, and avian retroviral integrases. J. Biol. Chem. 2001, 276, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Harper, A.L.; Sudol, M.; Katzman, M. An Amino Acid in the Central Catalytic Domain of Three Retroviral Integrases That Affects Target Site Selection in Nonviral DNA. J. Virol. 2003, 77, 3838–3845. [Google Scholar] [CrossRef] [Green Version]
- Enssle, J.; Mauer, B.; Schweizer, M.; Heinkelein, M.; Neumann-Haefelin, D.; Rethwilm, A.; Panhuysen, M.; Moebes, A. An active foamy virus integrase is required for virus replication. J. Gen. Virol. 1999, 80, 1445–1452. [Google Scholar] [CrossRef]
- Juretzek, T.; Holm, T.; Gärtner, K.; Kanzler, S.; Lindemann, D.; Herchenröder, O.; Picard-Maureau, M.; Rammling, M.; Heinkelein, M.; Rethwilm, A. Foamy Virus Integration. J. Virol. 2004, 78, 2472–2477. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, T.; Craigie, R. Integration of mini-retroviral DNA: A cell-free reaction for biochemical analysis of retroviral integration. Proc. Natl. Acad. Sci. USA 1989, 86, 3065–3069. [Google Scholar] [CrossRef]
- Katzman, M.; Katz, R.A.; Skalka, A.M.; Leis, J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 1989, 63, 5319–5327. [Google Scholar]
- Vora, A.C.; Fitzgerald, M.L.; Grandgenett, D.P. Removal of 3′-OH-terminal nucleotides from blunt-ended long terminal repeat termini by the avian retrovirus integration protein. J. Virol. 1990, 64, 5656–5659. [Google Scholar] [PubMed]
- Shin, C.-G. Characterization of Biochemical Properties of Feline Foamy Virus Integrase. J. Microbiol. Biotechnol. 2010, 20, 968–973. [Google Scholar] [CrossRef] [Green Version]
- Hazuda, D.J.; Felock, P.J.; Hastings, J.C.; Pramanik, B.; Wolfe, A.L. Differential divalent cation requirements uncouple the assembly and catalytic reactions of human immunodeficiency virus type 1 integrase. J. Virol. 1997, 71, 7005–7011. [Google Scholar] [PubMed]
- Mackler, R.; Lopez, M.; Osterhage, M.; Yoder, K. Prototype foamy virus integrase is promiscuous for target choice. Biochem. Biophys. Res. Commun. 2018, 503, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.D.; Lopez, M.A., Jr.; Hanne, J.; Peake, M.B.; Lee, J.-B.; Fishel, R.; Yoder, K.E. Retroviral intasomes search for a target DNA by 1D diffusion which rarely results in integration. Nat. Commun. 2016, 7, 11409. [Google Scholar] [CrossRef]
- Engelman, A.; Craigie, R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 1992, 66, 6361–6369. [Google Scholar] [PubMed]
- Cai, M.; Zheng, R.; Caffrey, M.; Craigie, R.; Clore, G.M.; Gronenborn, A.M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Genet. 1997, 4, 567–577. [Google Scholar] [CrossRef]
- Eijkelenboom, A.P.; Ent, F.M.V.D.; Vos, A.; Doreleijers, J.F.; Hård, K.; Tullius, T.D.; Plasterk, R.H.; Kaptein, R.; Boelens, R. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: A three-helix bundle stabilized by zinc. Curr. Biol. 1997, 7, 739–746. [Google Scholar] [CrossRef]
- Eijkelenboom, A.P.; Lutzke, R.A.P.; Boelens, R.; Plasterk, R.H.; Kaptein, R.; Hård, K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat. Struct. Mol. Biol. 1995, 2, 807–810. [Google Scholar] [CrossRef]
- Lodi, P.J.; Ernst, J.A.; Kuszewski, J.; Hickman, A.B.; Engelman, A.; Craigie, R.; Clore, G.M.; Gronenborn, A.M. Solution Structure of the DNA Binding Domain of HIV-1 Integrase. Biochemistry 1995, 34, 9826–9833. [Google Scholar] [CrossRef]
- Dyda, F.; Hickman, A.B.; Jenkins, T.M.; Engelman, A.; Craigie, R.; Davies, D.R. Crystal structure of the catalytic domain of HIV-1 integrase: Similarity to other polynucleotidyl transferases. Science 1994, 266, 1981–1986. [Google Scholar] [CrossRef]
- Nowotny, M. Retroviral integrase superfamily: The structural perspective. EMBO Rep. 2009, 10, 144–151. [Google Scholar] [CrossRef]
- Augé-Gouillou, C.; Brillet, B.; Hamelin, M.-H.; Bigot, Y. Assembly of the mariner Mos1 Synaptic Complex. Mol. Cell. Biol. 2005, 25, 2861–2870. [Google Scholar] [CrossRef] [PubMed]
- Hickman, A.B.; Perez, Z.N.; Zhou, L.Q.; Musingarimi, P.; Ghirlando, R.; Hinshaw, J.E.; Craig, N.L.; Dyda, F. Molecular architecture of a eukaryotic DNA transposase. Nat. Struct. Mol. Biol. 2005, 12, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Hickman, A.B.; Ewis, H.E.; Li, X.; Knapp, J.A.; Laver, T.; Doss, A.-L.; Tolun, G.; Steven, A.C.; Grishaev, A.; Bax, A.; et al. Structural Basis of hAT Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca domestica. Cell 2014, 158, 353–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, K.; Curtis, J.E.; Krueger, S.; Hwang, Y.; Cherepanov, P.; Bushman, F.D.; Van Duyne, G.D. Solution Conformations of Prototype Foamy Virus Integrase and its Stable Synaptic Complex with U5 Viral DNA. Structure 2012, 20, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lin, S.; Craigie, R. Outer domains of integrase within retroviral intasomes are dispensible for catalysis of DNA integration. Protein Sci. 2016, 25, 472–478. [Google Scholar] [CrossRef]
- Yin, Z.; Shi, K.; Banerjee, S.; Aihara, H. Crystal structure of the Rous sarcoma virus intasome. Crystal structure of the Rous sarcoma virus intasome. Nature 2016, 530, 362–366. [Google Scholar] [CrossRef]
- Allison Ballandras-Colas, M.B. Cryo-EM reveals a novel octameric integrase structure for β-retroviral intasome function. Nature 2016, 530, 358–361. [Google Scholar] [CrossRef]
- Ballandras-Colas, A.; Maskell, D.P.; Serrao, E.; Locke, J.; Swuec, P.; Jónsson, S.R.; Kotecha, A.; Cook, N.J.; Pye, V.E.; Taylor, I.A.; et al. A supramolecular assembly mediates lentiviral DNA integration. Science 2017, 355, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Passos, D.O.; Li, M.; Yang, R.; Rebensburg, S.V.; Ghirlando, R.; Jeon, Y.; Shkriabai, N.; Kvaratskhelia, M.; Craigie, R.; Lyumkis, D. CryoEM Structures and Atomic Model of the HIV-1 Strand Transfer Complex Intasome. Science 2017, 355, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Engelman, A.N.; Cherepanov, P. Retroviral intasomes arising. Curr. Opin. Struct. Biol. 2017, 47, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.C.-H.; Krucinski, J.; Miercke, L.J.W.; Finer-Moore, J.S.; Tang, A.H.; Leavitt, A.D.; Stroud, R.M. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: A model for viral DNA binding. Proc. Natl. Acad. Sci. USA 2000, 97, 8233–8238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.-G.; Su, S.-Y.; Robinson, H.; Padmanabhan, S.; Lim, L.; McCrary, B.S.; Edmondson, S.P.; Shriver, J.W.; Wang, A.H.-J. The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nat. Genet. 1998, 5, 782–786. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Jurado, K.A.; Lin, S.; Engelman, A.; Craigie, R. Engineered Hyperactive Integrase for Concerted HIV-1 DNA Integration. PLoS ONE 2014, 9, e105078. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.; Maertens, G.N.; Cherepanov, P. 3′-Processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 2012, 31, 3020–3028. [Google Scholar] [CrossRef]
- Serrao, E.; Krishnan, L.; Shun, M.-C.; Li, X.; Cherepanov, P.; Engelman, A.; Maertens, G.N. Integrase residues that determine nucleotide preferences at sites of HIV-1 integration: Implications for the mechanism of target DNA binding. Nucleic Acids Res. 2014, 42, 5164–5176. [Google Scholar] [CrossRef] [PubMed]
- Serrao, E.; Ballandras-Colas, A.; Cherepanov, P.; Maertens, G.N.; Engelman, A.N. Key determinants of target DNA recognition by retroviral intasomes. Retrovirology 2015, 12, 1295. [Google Scholar] [CrossRef] [PubMed]
- Naughtin, M.; Haftek-Terreau, Z.; Xavier, J.; Meyer, S.; Silvain, M.; Jaszczyszyn, Y.; Levy, N.; Miele, V.; Benleulmi, M.S.; Ruff, M.; et al. DNA Physical Properties and Nucleosome Positions Are Major Determinants of HIV-1 Integrase Selectivity. PLoS ONE 2015, 10, e0129427. [Google Scholar] [CrossRef]
- Benleulmi, M.S.; Matysiak, J.; Henriquez, D.R.; Vaillant, C.; Lesbats, P.; Calmels, C.; Naughtin, M.; Leon, O.; Skalka, A.M.; Ruff, M.; et al. Intasome architecture and chromatin density modulate retroviral integration into nucleosome. Retrovirology 2015, 12, 469. [Google Scholar] [CrossRef]
- Mackler, R.M.; Jones, N.D.; Gardner, A.M.; Lopez, M.A.; Howard, C.J.; Fishel, R.; Yoder, K.E. Nucleosome DNA unwrapping does not affect prototype foamy virus integration efficiency or site selection. PLoS ONE 2019, 14, e0212764. [Google Scholar] [CrossRef]
- Nowotny, M.; Yang, W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006, 25, 1924–1933. [Google Scholar] [CrossRef] [PubMed]
- Gangadharan, S.; Mularoni, L.; Fain-Thornton, J.; Wheelan, S.J.; Craig, N.L. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 21966–21972. [Google Scholar] [CrossRef] [PubMed]
- Montano, S.P.; Pigli, Y.Z.; Rice, P.A. The mu transpososome structure sheds light on DDE recombinase evolution. Nature 2012, 491, 413–417. [Google Scholar] [CrossRef]
- Pribil, P.A.; Haniford, D.B. Target DNA Bending is an Important Specificity Determinant in Target Site Selection in Tn10 Transposition. J. Mol. Biol. 2003, 330, 247–259. [Google Scholar] [CrossRef]
- Hallet, B.; Rezsöhazy, R.; Mahilton, J.; Delcour, J. IS231A insertion specificity: Consensus sequence and DNA bending at the target site. Mol. Microbiol. 1994, 14, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Rawal, R.K.; Rath, G.; Goyal, A.K. Structure Based Drug Design: Clinically Relevant HIV-1 Integrase Inhibitors. Curr. Top. Med. Chem. 2018, 18, 2664–2680. [Google Scholar] [CrossRef] [PubMed]
- Hazuda, D.J.; Hastings, J.C.; Wolfe, A.L.; Emini, E.A. A novel assay for the DNA strand-transfer reaction of HIV-1 integrase. Nucleic Acids Res. 1994, 22, 1121–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazuda, D.; Felock, P.J.; Hastings, J.C.; Pramanik, B.; Wolfe, A.L. Discovery and analysis of inhibitors of the human immunodeficiency integrase. Drug Des. Discov. 1997, 15, 17–24. [Google Scholar] [PubMed]
- Hazuda, D.J.; Felock, P.; Witmer, M.; Wolfe, A.; Stillmock, K.; Grobler, J.A.; Espeseth, A.; Gabryelski, L.; Schleif, W.; Blau, C.; et al. Inhibitors of Strand Transfer That Prevent Integration and Inhibit HIV-1 Replication in Cells. Science 2000, 287, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Marchand, C.; Burke, T.R.; Pommier, Y.; Nicklaus, M.C. Authentic HIV-1 integrase inhibitors. Futur. Med. Chem. 2010, 2, 1107–1122. [Google Scholar] [CrossRef] [Green Version]
- Cocohoba, J.; Dong, B.J. Raltegravir: The first HIV integrase inhibitor. Clin. Ther. 2008, 30, 1747–1765. [Google Scholar] [CrossRef]
- Sato, M.; Motomura, T.; Aramaki, H.; Matsuda, T.; Yamashita, M.; Ito, Y.; Kawakami, H.; Matsuzaki, Y.; Watanabe, W.; Yamataka, K.; et al. Novel HIV-1 Integrase Inhibitors Derived from Quinolone Antibiotics. J. Med. Chem. 2006, 49, 1506–1508. [Google Scholar] [CrossRef]
- Johns, B.A.; Kawasuji, T.; Weatherhead, J.G.; Taishi, T.; Temelkoff, D.P.; Yoshida, H.; Akiyama, T.; Taoda, Y.; Murai, H.; Kiyama, R.; et al. Carbamoyl pyridone HIV-1 integrase inhibitors 3. A diastereomeric approach to chiral nonracemic tricyclic ring systems and the discovery of dolutegravir (S/GSK1349572) and (S/GSK1265744). J. Med. Chem. 2013, 56, 5901–5916. [Google Scholar] [CrossRef]
- Barreca, M.L.; Ferro, S.; Rao, A.; De Luca, L.; Zappala’, M.; Monforte, A.-M.; Debyser, Z.; Witvrouw, M.; Chimirri, A. Pharmacophore-Based Design of HIV-1 Integrase Strand-Transfer Inhibitors. J. Med. Chem. 2005, 48, 7084–7088. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Rong, J.; Zhang, B.; Hu, L.; Wang, X.; Zeng, C. Design and synthesis of N-methylpyrimidone derivatives as HIV-1 integrase inhibitors. Bioorg. Med. Chem. 2015, 23, 735–741. [Google Scholar] [CrossRef]
- Cuzzucoli Crucitti, G.; Métifiot, M.; Pescatori, L.; Messore, A.; Madia, V.N.; Pupo, G.; Saccoliti, F.; Scipione, L.; Tortorella, S.; Esposito, F.; et al. Structure-activity relationship of pyrrolyl diketo acid derivatives as dual inhibitors of HIV-1 integrase and reverse transcriptase ribonuclease H domain. J. Med. Chem. 2015, 58, 1915–1928. [Google Scholar] [CrossRef]
- Yoshinaga, T.; Kobayashi, M.; Seki, T.; Miki, S.; Wakasa-Morimoto, C.; Suyama-Kagitani, A.; Kawauchi-Miki, S.; Taishi, T.; Kawasuji, T.; Johns, B.A.; et al. Antiviral characteristics of GSK1265744, an HIV integrase inhibitor dosed orally or by long-acting injection. Antimicrob. Agents Chemother. 2015, 59, 397–406. [Google Scholar] [CrossRef]
- Tsiang, M.; Jones, G.S.; Goldsmith, J.; Mulato, A.; Hansen, D.; Kan, E.; Tsai, L.; Bam, R.A.; Stepan, G.; Stray, K.M.; et al. Antiviral Activity of Bictegravir (GS-9883), a Novel Potent HIV-1 Integrase Strand Transfer Inhibitor with an Improved Resistance Profile. Antimicrob. Agents Chemother. 2016, 60, 7086–7097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trezza, C.; Ford, S.L.; Spreen, W.; Pan, R.; Piscitelli, S. Formulation and pharmacology of long-acting cabotegravir. Curr. Opin. HIV AIDS 2015, 10, 239–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzkee, N.C.; Masse, J.E.; Shen, Y.; Davies, D.R.; Bax, A. Solution Conformation and Dynamics of the HIV-1 Integrase Core Domain. J. Biol. Chem. 2010, 285, 18072–18084. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, L.; Li, X.; Naraharisetty, H.L.; Hare, S.; Cherepanov, P.; Engelman, A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. USA 2010, 107, 15910–15915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, B.C.; Métifiot, M.; Pommier, Y.; Hughes, S.H. Molecular dynamics approaches estimate the binding energy of HIV-1 integrase inhibitors and correlate with in vitro activity. Antimicrob. Agents Chemother. 2012, 56, 411–419. [Google Scholar] [CrossRef]
- Hu, J.-P.; He, H.-Q.; Tang, D.-Y.; Sun, G.-F.; Zhang, Y.-Q.; Fan, J.; Chang, S. Study on the interactions between diketo-acid inhibitors and prototype foamy virus integrase-DNA complex via molecular docking and comparative molecular dynamics simulation methods. J. Biomol. Struct. Dyn. 2013, 31, 734–747. [Google Scholar] [CrossRef]
- Goethals, O.; Van Ginderen, M.; Vos, A.; Cummings, M.D.; Van Der Borght, K.; Van Wesenbeeck, L.; Feyaerts, M.; Verheyen, A.; Smits, V.; Van Loock, M.; et al. Resistance to raltegravir highlights integrase mutations at codon 148 in conferring cross-resistance to a second-generation HIV-1 integrase inhibitor. Antivir. Res. 2011, 91, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Hare, S.; Vos, A.M.; Clayton, R.F.; Thuring, J.W.; Cummings, M.D.; Cherepanov, P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc. Natl. Acad. Sci. USA 2010, 107, 20057–20062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hare, S.; Smith, S.J.; Métifiot, M.; Jaxa-Chamiec, A.; Pommier, Y.; Hughes, S.H.; Cherepanov, P. Structural and Functional Analyses of the Second-Generation Integrase Strand Transfer Inhibitor Dolutegravir (S/GSK1349572). Mol. Pharmacol. 2011, 80, 565–572. [Google Scholar] [CrossRef] [Green Version]
- Johnson, B.C.; Metifiot, M.; Ferris, A.; Pommier, Y.; Hughes, S.H. A Homology Model of HIV-1 Integrase and Analysis of Mutations Designed to Test the Model. J. Mol. Biol. 2013, 425, 2133–2146. [Google Scholar] [CrossRef]
- Hu, J.; Liu, M.; Tang, D.; Chang, S. Substrate Recognition and Motion Mode Analyses of PFV Integrase in Complex with Viral DNA via Coarse-Grained Models. PLoS ONE 2013, 8, e54929. [Google Scholar] [CrossRef]
- Du, W.; Zuo, K.; Sun, X.; Liu, W.; Yan, X.; Liang, L.; Wan, H.; Chen, F.; Hu, J. An effective HIV-1 integrase inhibitor screening platform: Rationality validation of drug screening, conformational mobility and molecular recognition analysis for PFV integrase complex with viral DNA. J. Mol. Graph. Model. 2017, 78, 96–109. [Google Scholar] [CrossRef]
- Quashie, P.K.; Han, Y.-S.; Hassounah, S.; Mesplède, T.; Wainberg, M.A. Structural Studies of the HIV-1 Integrase Protein: Compound Screening and Characterization of a DNA-Binding Inhibitor. PLoS ONE 2015, 10, e0128310. [Google Scholar] [CrossRef]
- Reddy, K.K.; Singh, S.K. Combined ligand and structure-based approaches on HIV-1 integrase strand transfer inhibitors. Chem. Interact. 2014, 218, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Dayer, M.R. Comparison of Newly Assembled Full Length HIV-1 Integrase With Prototype Foamy Virus Integrase: Structure-Function Prospective. Jundishapur J. Microbiol. 2016, 9, e29773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benleulmi, M.S.; Matysiak, J.; Robert, X.; Miskey, C.; Mauro, E.; Lapaillerie, D.; Lesbats, P.; Chaignepain, S.; Henriquez, D.R.; Calmels, C.; et al. Modulation of the functional association between the HIV-1 intasome and the nucleosome by histone amino-terminal tails. Retrovirology 2017, 14, 54. [Google Scholar] [CrossRef] [Green Version]
- Mauro, E.; Lesbats, P.; Lapaillerie, D.; Chaignepain, S.; Maillot, B.; Oladosu, O.; Robert, X.; Fiorini, F.; Kieffer, B.; Bouaziz, S.; et al. Human H4 tail stimulates HIV-1 integration through binding to the carboxy-terminal domain of integrase. Nucleic Acids Res. 2019, 47, 3607–3618. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.Z.; Wang, Y.; Goff, S.P. Histones are rapidly loaded onto unintegrated retroviral DNAs soon after nuclear entry. Cell Host Microbe 2016, 20, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, G.Z.; Cingöz, O.; Goff, S.P. NP220 mediates silencing of unintegrated retroviral DNA. Nature 2018, 564, 278–282. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, G.-E.; Mauro, E.; Parissi, V.; Shin, C.-G.; Lesbats, P. Structural Insights on Retroviral DNA Integration: Learning from Foamy Viruses. Viruses 2019, 11, 770. https://doi.org/10.3390/v11090770
Lee G-E, Mauro E, Parissi V, Shin C-G, Lesbats P. Structural Insights on Retroviral DNA Integration: Learning from Foamy Viruses. Viruses. 2019; 11(9):770. https://doi.org/10.3390/v11090770
Chicago/Turabian StyleLee, Ga-Eun, Eric Mauro, Vincent Parissi, Cha-Gyun Shin, and Paul Lesbats. 2019. "Structural Insights on Retroviral DNA Integration: Learning from Foamy Viruses" Viruses 11, no. 9: 770. https://doi.org/10.3390/v11090770




