RNA Back and Forth: Looking through Ribozyme and Viroid Motifs
Abstract
:1. Introduction
1.1. The RNA World
1.2. Viroids and Ribozymes
1.3. RNA Dimerization: Bonded/Non-Bonded Dimers
1.3.1. Bonded Dimers or Double-Hammerhead Structures
1.3.2. Non-Bonded Dimers
1.4. Small Angle Neutron Scattering (SANS) and the Study of Multimeric Complexes
1.5. Viroids in Non-Plant Hosts
2. ASBVd as a Viroid Model
2.1. ASBVd (+)/(−) and the Biocatalyst/Template Paradox
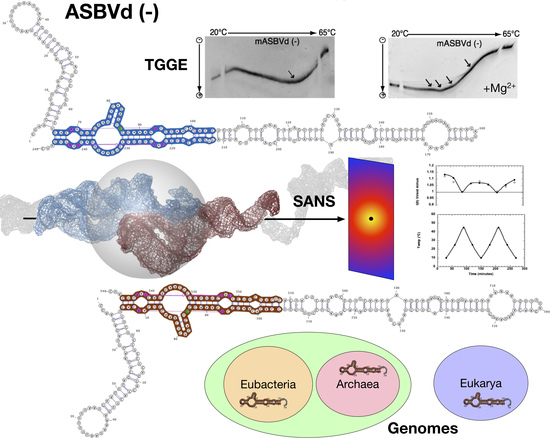
2.2. Viroid Structures and Interactions Revealed by SANS
2.3. Replication of ASBVd in Other Kingdoms
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ASBVd | Avocado Sun Blotch Viroid |
| mASBVd | monomeric ASBVd |
| pRLASBVd | plasmid recombinant ASBVd derived from the pRL25 plasmid |
| HHR | Hammerhead ribozyme |
| TGGE | Temperature gradient gel electrophoresis |
| ADHR | adenine-dependent hairpin ribozyme |
| SANS | small-angle neutron scattering |
References
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Woese, C.R. On the evolution of the genetic code. Proc. Natl. Acad. Sci. USA 1965, 54, 1546–1552. [Google Scholar] [CrossRef] [PubMed]
- Crick, F. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Orgel, L. Evolution of the genetic apparatus. J. Mol. Biol. 1968, 38, 381–393. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Guerrier-Takada, C.; Altman, S. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science 1984, 223, 285–286. [Google Scholar] [CrossRef]
- Bajaj, P.; Steger, G.; Hammann, C. Sequence elements outside the catalytic core of natural hairpin ribozymes modulate the reactions differentially. Biol. Chem. 2011, 392, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Serra, P.; Minoia, S.; Serio, F.D.; Navarro, B. Viroids: From Genotype to Phenotype Just Relying on RNA Sequence and Structural Motifs. Front. Microbiol. 2012, 3, 217. [Google Scholar] [CrossRef]
- Bajaj, P.; Hammann, C. Characterization of Hairpin Ribozyme Reactions. In Therapeutic Applications of Ribozymes and Riboswitches: Methods and Protocols; Lafontaine, D., Dubé, A., Eds.; Humana Press: Totowa, NJ, USA, 2014; pp. 97–111. [Google Scholar]
- Jimenez, R.M.; Polanco, J.A.; Lupták, A. Chemistry and Biology of Self-Cleaving Ribozymes. Trends Biochem. Sci. 2015, 40, 648–661. [Google Scholar] [CrossRef]
- Salehi-Ashtiani, K.; Lupták, A.; Litovchick, A.; Szostak, J.W. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 2006, 313, 1788–1792. [Google Scholar] [CrossRef]
- Webb, C.H.T.; Riccitelli, N.J.; Ruminski, D.J.; Lupták, A. Widespread occurrence of self-cleaving ribozymes. Science 2009, 326, 953. [Google Scholar] [CrossRef]
- Webb, C.H.T.; Lupták, A. HDV-like self-cleaving ribozymes. RNA Biol. 2011, 8, 719–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Luque, F.J.; López, M.C.; Carreira, P.E.; Alonso, C.; Thomas, M.C. The wide expansion of hepatitis delta virus-like ribozymes throughout trypanosomatid genomes is linked to the spreading of L1Tc/ingi clade mobile elements. BMC Genom. 2014, 15, 340. [Google Scholar] [CrossRef] [PubMed]
- Brazas, R.; Ganem, D. A cellular homolog of hepatitis delta antigen: Implications for viral replication and evolution. Science 1996, 274, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.D. How did replicating and coding RNAs first get together? Science 1996, 274, 66–67. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Pelchat, M. Origin of hepatitis delta virus. Future Microbiol. 2010, 5, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Ruiz-Ruiz, S.; Serra, P. Viroids and hepatitis delta virus. Semin. Liver Dis. 2012, 32, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013, 3, 546–557. [Google Scholar] [CrossRef] [Green Version]
- Lempp, F.A.; Ni, Y.; Urban, S. Hepatitis delta virus: Insights into a peculiar pathogen and novel treatment options. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 580–589. [Google Scholar] [CrossRef]
- Flores, R.; Owens, R.A.; Taylor, J. Pathogenesis by subviral agents: Viroids and hepatitis delta virus. Curr. Opin. Virol. 2016, 17, 87–94. [Google Scholar] [CrossRef]
- Lünse, C.E.; Weinberg, Z.; Breaker, R.R. Numerous small hammerhead ribozyme variants associated with Penelope-like retrotransposons cleave RNA as dimers. RNA Biol. 2016, 14, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Cervera, A.; Urbina, D.; de la Peña, M. Retrozymes are a unique family of non-autonomous retro transposons with hammerhead ribo zymes that propagate in plants through circular RNAs. Genome Biol. 2016, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- De la Peña, M.; García-Robles, I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA 2010, 16, 1943–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Peña, M.; García-Robles, I. Intronic hammerhead ribozymes are ultraconserved in the human genome. EMBO Rep. 2010, 11, 711–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seehafer, C.; Kalweit, A.; Steger, G.; Gräf, S.; Hammann, C. From alpaca to zebrafish: Hammerhead ribozymes wherever you look. RNA 2010, 17, 21–26. [Google Scholar] [CrossRef] [Green Version]
- Jimenez, R.M.; Delwart, E.; Lupták, A. Structure-based search reveals hammerhead ribozymes in the human microbiome. J. Biol. Chem. 2011, 286, 7737–7743. [Google Scholar] [CrossRef]
- Perreault, J.; Weinberg, Z.; Roth, A.; Popescu, O.; Chartrand, P.; Ferbeyre, G.; Breaker, R.R. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput. Biol. 2011, 7, e1002031. [Google Scholar] [CrossRef]
- Senčilo, A.; Jacobs-Sera, D.; Russell, D.A.; Ko, C.C.; Bowman, C.A.; Atanasova, N.S.; Österlund, E.; Oksanen, H.M.; Bamford, D.H.; Hatfull, G.F.; Roine, E.; Hendrix, R.W. Snapshot of haloarchaeal tailed virus genomes. RNA Biol. 2013, 10, 803–816. [Google Scholar] [CrossRef]
- Wurmthaler, L.A.; Klauser, B.; Hartig, J.S. Highly motif- and organism-dependent effects of naturally occurring hammerhead ribozyme sequences on gene expression. RNA Biol. 2018, 15, 231–241. [Google Scholar] [CrossRef]
- Cervera, A.; de la Peña, M. Eukaryotic penelope-like retroelements encode hammerhead ribozyme motifs. Mol. Biol. Evol. 2014, 31, 2941–2947. [Google Scholar] [CrossRef]
- Suslov, N.B.; DasGupta, S.; Huang, H.; Fuller, J.R.; Lilley, D.M.J.; Rice, P.A.; Piccirilli, J.A. Crystal structure of the Varkud satellite ribozyme. Nat. Chem. Biol. 2015, 11, 840–846. [Google Scholar] [CrossRef]
- Leclerc, F.; Zaccai, G.; Vergne, J.; Řìhovà, M.; Martel, A.; Maurel, M.C. Self-assembly Controls Self-cleavage of HHR from ASBVd (−): A Combined SANS and Modeling Study. Sci. Rep. 2016, 6, 30287. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Maurel, M.; Ebel, C.; Vergne, J.; Pipich, V.; Zaccai, G. Self-association of adenine-dependent hairpin ribozymes. Eur. Biophys. J. 2008, 37, 173–182. [Google Scholar] [CrossRef]
- Tanaka, T.; Matsumura, S.; Furuta, H.; Ikawa, Y. Tecto-GIRz: Engineered Group I Ribozyme the Catalytic Ability of Which Can Be Controlled by Self-Dimerization. ChemBioChem Eur. J. Chem. Biol. 2016, 17, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Diener, T.O. Circular RNAs: Relics of precellular evolution? Proc. Natl. Acad. Sci. USA 1989, 86, 9370–9374. [Google Scholar] [CrossRef]
- Flores, R.; Gago-Zachert, S.; Serra, P.; Sanjuán, R.; Elena, S.F. Viroids: Survivors from the RNA world? Microbiology 2013, 68, 395–414. [Google Scholar] [CrossRef] [PubMed]
- Diener, T.O. Viroids: “living fossils” of primordial RNAs? Biol. Direct. 2016, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Diener, T. Potato spindle tuber “virus”. Virology 1971, 45, 411–428. [Google Scholar] [CrossRef]
- Ohnishi, K.; Hokari, S.; Shutou, H.; Ohshima, M.; Furuichi, N.; Goda, M. Origin of most primitive mRNAs and genetic codes via interactions between primitive tRNA ribozymes. Genome Inform. 2002, 13, 71–81. [Google Scholar]
- Ohnishi, K.; Ohshima, M.; Furuichi, N. Evolution from possible primitive tRNA-viroids to early poly-tRNA-derived mRNAs: A new approach from the poly-tRNA theory. Genome Inform. 2005, 16, 94–103. [Google Scholar]
- Hernández, C.; Daròs, J.; Elena, S.; Moya, A.; Flores, R. The strands of both polarities of a small circular RNA from carnation self-cleave in vitro through alternative double- and single-hammerhead structures. Nucleic Acids Res. 1992, 20, 6323–6329. [Google Scholar] [CrossRef] [Green Version]
- De la Peña, M.; Cervera, A. Circular RNAs with hammerhead ribozymes encoded in eukaryotic genomes: The enemy at home. RNA Biol. 2017, 14, 985–991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daros, J.A.; Marcos, J.F.; Hernandez, C.; Flores, R. Replication of avocado sunblotch viroid: Evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc. Natl. Acad. Sci. USA 1994, 91, 12813–12817. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Daròs, J.; Hernández, C. Avsunviroidae family: Viroids containing hammerhead ribozymes. Adv. Virus Res. 2000, 55, 271–323. [Google Scholar] [PubMed]
- Flores, R.; Gas, M.E.; Molina-Serrano, D.; Nohales, M.Á.; Carbonell, A.; Gago, S.; la Peña, M.D.; Daròs, J.A. Viroid Replication: Rolling-Circles Enzymes and Ribozymes. Viruses 2009, 1, 317–334. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, F. Hammerhead Ribozymes: True Metal or Nucleobase Catalysis? Where Is the Catalytic Power from? Molecules 2010, 15, 5389–5407. [Google Scholar] [CrossRef] [Green Version]
- Mir, A.; Chen, J.; Robinson, K.; Lendy, E.; Goodman, J.; Neau, D.; Golden, B.L. Two Divalent Metal Ions and Conformational Changes Play Roles in the Hammerhead Ribozyme Cleavage Reaction. Biochemistry 2015, 54, 6369–6381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mir, A.; Golden, B.L. Two Active Site Divalent Ions in the Crystal Structure of the Hammerhead Ribozyme Bound to a Transition State Analogue. Biochemistry 2016, 55, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Giese, T.J.; Golden, B.L.; York, D.M. Divalent Metal Ion Activation of a Guanine General Base in the Hammerhead Ribozyme: Insights from Molecular Simulations. Biochemistry 2017, 56, 2985–2994. [Google Scholar] [CrossRef] [PubMed]
- Canny, M.D.; Jucker, F.M.; Pardi, A. Efficient Ligation of theSchistosomaHammerhead Ribozyme. Biochemistry 2007, 46, 3826–3834. [Google Scholar] [CrossRef] [PubMed]
- Ivica, N.A.; Obermayer, B.; Campbell, G.W.; Rajamani, S.; Gerland, U.; Chen, I.A. The paradox of dual roles in the RNA world: Resolving the conflict between stable folding and templating ability. J. Mol. Evol. 2013, 77, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Forster, A.C.; Davies, C.; Sheldon, C.C.; Jeffries, A.C.; Symons, R.H. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature 1988, 334, 265–267. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.F.; Gellatly, D.L.; Sehgal, O.P.; Abouhaidar, M.G. Self-cleaving circular RNA associated with rice yellow mottle virus is the smallest viroid-like RNA. Virology 1998, 241, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Song, S.I.; Silver, S.L.; Aulik, M.A.; Rasochova, L.; Mohan, B.R.; Miller, W.A. Satellite cereal yellow dwarf virus-RPV (satRPV) RNA requires a douXble hammerhead for self-cleavage and an alternative structure for replication. J. Mol. Biol. 1999, 293, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Beaudry, D.; Busière, F.; Lareau, F.; Lessard, C.; Perreault, J.P. The RNA of both polarities of the peach latent mosaic viroid self-cleaves in vitro solely by single hammerhead structures. Nucleic Acids Res. 1995, 23, 745–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.I.; Miller, W.A. Cis and trans requirements for rolling circle replication of a satellite RNA. J. Virol. 2004, 78, 3072–3082. [Google Scholar] [CrossRef] [PubMed]
- Daròs, J.A. Eggplant latent viroid: A friendly experimental system in the family Avsunviroidae. Mol. Plant Pathol. 2016, 17, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Serra, P.; Messmer, A.; Sanderson, D.; James, D.; Flores, R. Apple hammerhead viroid-like RNA is a bona fide viroid: Autonomous replication and structural features support its inclusion as a new member in the genus Pelamoviroid. Virus Res. 2018, 249, 8–15. [Google Scholar] [CrossRef]
- Davies, C.; Sheldon, C.C.; Symons, R.H. Alternative hammerhead structures in the self-cleavage of avocado sunblotch viroid RNAs. Nucleic Acids Res. 1991, 19, 1893–1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biondi, E.; Branciamore, S.; Fusi, L.; Gago, S.; Gallori, E. Catalytic activity of hammerhead ribozymes in a clay mineral environment: Implications for the RNA world. Gene 2007, 389, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Heinicke, L.A.; Bevilacqua, P.C. Activation of PKR by RNA misfolding: HDV ribozyme dimers activate PKR. RNA 2012, 18, 2157–2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaccai, N.R.; Serdyuk, I.N.; Zaccai, J. Methods in Molecular Biophysics: Structure, Dynamics, Function for Biology and Medicine, 2nd ed.; Cambridge University Press: Cambridge, UK, 2017. [Google Scholar]
- Bernadó, P.; Shimizu, N.; Zaccai, G.; Kamikubo, H.; Sugiyama, M. Solution scattering approaches to dynamical ordering in biomolecular systems. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 253–274. [Google Scholar] [CrossRef] [PubMed]
- Przybilski, R.; Gräf, S.; Lescoute, A.; Nellen, W.; Westhof, E.; Steger, G.; Hammann, C. Functional Hammerhead Ribozymes Naturally Encoded in the Genome of Arabidopsis thaliana. Plant Cell 2005, 17, Y1877–Y1885. [Google Scholar] [CrossRef] [PubMed]
- Pooggin, M.M. Small RNA-Omics for Plant Virus Identification, Virome Reconstruction, and Antiviral Defense Characterization. Front. Microbiol. 2018, 9, 2779. [Google Scholar] [CrossRef] [PubMed]
- Walia, Y.; Dhir, S.; Zaidi, A.A.; Hallan, V. Apple scar skin viroid naked RNA is actively transmitted by the whitefly Trialeurodes vaporariorum. RNA Biol. 2015, 12, 1131–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchins, C.; Keese, P.; Visvader, J.; Rathjen, P.; McInnes, J.; Symons, R. Comparison of multimeric plus and minus forms of viroids and virusoids. Plant Mol. Biol. 1985, 4, 293–304. [Google Scholar] [CrossRef]
- Riesner, D.; Steger, G. Temperature-Gradient Gel Electrophoresis of RNA. In Handbook of RNA Biochemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; Chapter 21; pp. 427–444. [Google Scholar]
- Delan-Forino, C.; Deforges, J.; Benard, L.; Sargueil, B.; Maurel, M.; Torchet, C. Structural analyses of Avocado sunblotch viroid reveal differences in the folding of plus and minus RNA strands. Viruses 2014, 6, 489–506. [Google Scholar] [CrossRef]
- Delan-Forino, C.; Maurel, M.C.; Torchet, C. Replication of Avocado Sunblotch Viroid in the Yeast Saccharomyces cerevisiae. J. Virol. 2011, 85, 3229–3238. [Google Scholar] [CrossRef] [PubMed]
- Sato, N. Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci. 2001, 6, 151–155. [Google Scholar] [CrossRef]
- Latifi, A.; Bernard, C.; da Silva, L.; Andéol, Y.; Elleuch, A.; Risoul, V.; Vergne, J.; Maurel, M.C. Replication of Avocado Sunblotch Viroid in the Cyanobacterium Nostoc Sp. PCC 7120. J. Plant Pathol. Microbiol. 2016, 7, 2. [Google Scholar] [CrossRef]
- Gago, S.; Elena, S.F.; Flores, R.; Sanjuán, R. Extremely high mutation rate of a hammerhead viroid. Science 2009, 323, 1308. [Google Scholar] [CrossRef] [PubMed]
- Elena, S.F.; Gomez, G.; Daròs, J.A. Evolutionary constraints to viroid evolution. Viruses 2009, 1, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Horning, D.P.; Joyce, G.F. Amplification of RNA by an RNA polymerase ribozyme. Proc. Natl. Acad. Sci. USA 2016, 113, 9786–9791. [Google Scholar] [CrossRef] [Green Version]
- Bergman, N.H. The three-dimensional architecture of the class I ligase ribozyme. RNA 2004, 10, 176–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leclerc, F.; Tisné, C. Des viroïdes et virus à l’homme, dans les traces du “RNA World”. In Contribution au Livre Blanc de l’Institut des Sciences de la Vie du CNRS; Jesus, C., Ed.; CNRS Editions: Paris, France, 2017. [Google Scholar]
- Semancik, J.S.; Szychowski, J.A. Avocado sunblotch disease: A persistent viroid infection in which variants are associated with differential symptoms. J. Gen. Virol. 1994, 75, 1543–1549. [Google Scholar] [CrossRef] [PubMed]
- Giguère, T.; Adkar-Purushothama, C.R.; Bolduc, F.; Perreault, J.P. Elucidation of the structures of all members of the Avsunviroidae family. Mol. Plant Pathol. 2014, 15, 767–779. [Google Scholar] [CrossRef]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maurel, M.-C.; Leclerc, F.; Vergne, J.; Zaccai, G. RNA Back and Forth: Looking through Ribozyme and Viroid Motifs. Viruses 2019, 11, 283. https://doi.org/10.3390/v11030283
Maurel M-C, Leclerc F, Vergne J, Zaccai G. RNA Back and Forth: Looking through Ribozyme and Viroid Motifs. Viruses. 2019; 11(3):283. https://doi.org/10.3390/v11030283
Chicago/Turabian StyleMaurel, Marie-Christine, Fabrice Leclerc, Jacques Vergne, and Giuseppe Zaccai. 2019. "RNA Back and Forth: Looking through Ribozyme and Viroid Motifs" Viruses 11, no. 3: 283. https://doi.org/10.3390/v11030283