Potato Virus Y Infection Alters Small RNA Metabolism and Immune Response in Tomato
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant and Virus Material
2.2. RNA Extraction
2.3. Primer Design and RT-qPCR Analysis for Detection of Viral RNA, mRNAs and miRNAs
2.4. Sequencing and sRNA Data Analysis
2.5. Identification of Differentially Expressed miRNAs
2.6. Target Gene Prediction and Functional Analysis
2.7. Degradome-Seq Target Validation
2.8. Validation of miRNA Targets with RLM 5´-RACE
2.9. Data Availability and Retrieval
3. Results
3.1. Tomato plants Exhibit Symptom Recovery at Later Stages of PVYC-to Infection
3.2. Viral siRNA Accumulation Levels Correlate with Viral RNA Titer
3.3. Known and Novel miRNAs and Their isomiR Variants Were Identified in Tomato Leaf Tissues
3.4. Regulation of Known and Novel Tomato mRNA upon PVYC-to Infection
3.5. RT-qPCR Expression Analysis of Known and Novel miRNAs and Their Target mRNAs
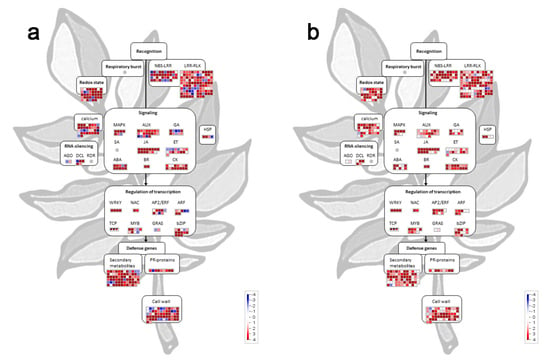
3.6. Target Genes Prediction and Functional Characterization of Differentially Expressed Tomato miRNAs
3.7. PVYC-to Infection Induces Secondary Phased siRNA Accumulation
3.8. miRNAs Mediate Cleavage of Transcripts Encoding NLRs and RLPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big impact on plant development. Trends Plant Sci. 2017, 22, 1056–1068. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef]
- Yang, L.; Huang, H. Roles of small RNAs in plant disease resistance. J. Integr. Plant Biol. 2014, 56, 962–970. [Google Scholar] [CrossRef]
- Fei, Q.; Zhang, Y.; Xia, R.; Meyers, B.C. Small RNAs Add Zing to the Zig-Zag-Zig Model of Plant Defenses. Mol. Plant-Microbe Interact. 2016, 29, 165–169. [Google Scholar] [CrossRef]
- Huang, J.; Yang, M.; Zhang, X. The function of small RNAs in plant biotic stress response. J. Integr. Plant Biol. 2016, 58, 312–327. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Y. Dissection of RNAi-based antiviral immunity in plants. Curr. Opin. Virol. 2018, 32, 88–99. [Google Scholar] [CrossRef]
- Meng, Y.; Shao, C.; Wang, H.; Chen, M. The Regulatory Activities of Plant MicroRNAs: A More Dynamic Perspective. Plant Physiol. 2011, 157, 1583–1595. [Google Scholar] [CrossRef]
- Bai, M.; Yang, G.S.; Chen, W.T.; Mao, Z.C.; Kang, H.X.; Chen, G.H.; Yang, Y.H.; Xie, B.Y. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 2012, 501, 52–62. [Google Scholar] [CrossRef]
- Wang, X.B.; Wu, Q.; Ito, T.; Cillo, F.; Li, W.X.; Chen, X.; Yu, J.L.; Ding, S.W. RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.; Muhammad, S.; Cao, M.; Wu, L. Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnol. J. 2018, 16, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Križnik, M.; Petek, M.; Dobnik, D.; Ramsak, Z.; Baebler, S.; Pollmann, S.; Kreuze, J.F.; Zel, J.; Gruden, K. Salicylic Acid Perturbs sRNA-Gibberellin Regulatory Network in Immune Response of Potato to Potato virus Y Infection. Front. Plant Sci. 2017, 8, 2192. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef]
- Ding, S.-W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Stavolone, L.; Prigigallo, M.I.; Cillo, F. Plant viruses against RNA silencing based defenses: Strategies and solutions. In Applied Plant Biotechnology for Improving Resistance to Biotic Stress; Palmiro, P., Yiguo, H., Eds.; Acadamic Press: New York, NY, USA, 2019; pp. 225–250. [Google Scholar] [CrossRef]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479, 85–103. [Google Scholar] [CrossRef]
- Kasschau, K.D.; Xie, Z.; Allen, E.; Llave, C.; Chapman, E.J.; Krizan, K.A.; Carrington, J.C. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 2003, 4, 205–217. [Google Scholar] [CrossRef]
- Lewsey, M.; Robertson, F.C.; Canto, T.; Palukaitis, P.; Carr, J.P. Selective targeting of miRNA-regulated plant development by a viral counter-silencing protein. Plant J. 2007, 50, 240–252. [Google Scholar] [CrossRef]
- Díaz-Pendón, J.A.; Ding, S.-W. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 2008, 46, 303–326. [Google Scholar] [CrossRef]
- Cillo, F.; Mascia, T.; Pasciuto, M.M.; Gallitelli, D. Differential effects of mild and severe Cucumber mosaic virus strains in the perturbation of MicroRNA-regulated gene expression in tomato map to the 3’ sequence of RNA 2. Mol. Plant Microbe Interact. 2009, 22, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Sanfacon, H. Symptom recovery in virus-infected plants: Revisiting the role of RNA silencing mechanisms. Virology 2015, 479, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Molen, T.A. Host recovery and reduced virus level in the upper leaves after Potato virus Y infection occur in tobacco and tomato but not in potato plants. Viruses 2015, 7, 680–698. [Google Scholar] [CrossRef] [PubMed]
- Scholthof, K.B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, J.H.; Meacham, T.; Berger, P.H.; Shiel, P.J.; Crosslin, J.M.; Hamm, P.B.; Kopp, H. Whole genome characterization of Potato virus Y isolates collected in the western USA and their comparison to isolates from Europe and Canada. Arch. Virol. 2006, 151, 1055–1074. [Google Scholar] [CrossRef]
- Ogawa, T.; Tomitaka, Y.; Nakagawa, A.; Ohshima, K. Genetic structure of a population of Potato virus Y inducing potato tuber necrotic ringspot disease in Japan; comparison with North American and European populations. Virus Res. 2008, 131, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Jia, M.A.; Yang, Y.; Zhan, L.; Cheng, X.; Cai, J.; Zhang, J.; Yang, J.; Liu, T.; Fu, Q.; et al. Integrated analysis of tobacco miRNA and mRNA expression profiles under PVY infection provids insight into tobacco-PVY interactions. Sci. Rep. 2017, 7, 4895. [Google Scholar] [CrossRef]
- Diermann, N.; Matousek, J.; Junge, M.; Riesner, D.; Steger, G. Characterization of plant miRNAs and small RNAs derived from potato spindle tuber viroid (PSTVd) in infected tomato. Biol. Chem. 2010, 391, 1379–1390. [Google Scholar] [CrossRef]
- Feng, J.; Liu, S.; Wang, M.; Lang, Q.; Jin, C. Identification of microRNAs and their targets in tomato infected with Cucumber mosaic virus based on deep sequencing. Planta 2014, 240, 1335–1352. [Google Scholar] [CrossRef]
- Pradhan, B.; Naqvi, A.R.; Saraf, S.; Mukherjee, S.K.; Dey, N. Prediction and characterization of Tomato leaf curl New Delhi virus (ToLCNDV) responsive novel microRNAs in Solanum lycopersicum. Virus Res. 2015, 195, 183–195. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Y.; Ding, B.; Fei, Z. Comprehensive Transcriptome Analyses Reveal that Potato Spindle Tuber Viroid Triggers Genome-Wide Changes in Alternative Splicing, Inducible trans-Acting Activity of Phased Secondary Small Interfering RNAs, and Immune Responses. J. Virol. 2017, 91, e00247-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, A.; Goswami, K.; Tiwari, M.; Mukherjee, S.K.; Sanan-Mishra, N. Identification and comparative analysis of microRNAs from tomato varieties showing contrasting response to ToLCV infections. Physiol. Mol. Biol. Plants 2018, 24, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Mascia, T.; Finetti-Sialer, M.; Cillo, F.; Gallitelli, D. Biological and molecular characterization of a recombinant isolate of potato virus Y associated with a tomato necrotic disease occurring in Italy. J. Plant Pathol. 2010, 92, 131–138. [Google Scholar]
- Mascia, T.; Santovito, E.; Gallitelli, D.; Cillo, F. Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Mol. Plant Pathol. 2010, 11, 805–816. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Moxon, S.; Schwach, F.; Dalmay, T.; Maclean, D.; Studholme, D.J.; Moulton, V. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 2008, 24, 2252–2253. [Google Scholar] [CrossRef] [Green Version]
- Bateman, A.; Agrawal, S.; Birney, E.; Bruford, E.A.; Bujnicki, J.M.; Cochrane, G.; Cole, J.R.; Dinger, M.E.; Enright, A.J.; Gardner, P.P.; et al. RNAcentral: A vision for an international database of RNA sequences. RNA 2011, 17, 1941–1946. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [Green Version]
- Shahid, S.; Axtell, M.J. Identification and annotation of small RNA genes using ShortStack. Methods 2014, 67, 20–27. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Sun, Y. miR-PREFeR: An accurate, fast and easy-to-use plant miRNA prediction tool using small RNA-Seq data. Bioinformatics 2014, 30, 2837–2839. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, L.F.V.; Christoff, A.P.; Margis, R. isomiRID: A framework to identify microRNA isoforms. Bioinformatics 2013, 29, 2521–2523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neilsen, C.T.; Goodall, G.J.; Bracken, C.P. IsomiRs—the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012, 28, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuperus, J.T.; Carbonell, A.; Fahlgren, N.; Garcia-Ruiz, H.; Burke, R.T.; Takeda, A.; Sullivan, C.M.; Gilbert, S.D.; Montgomery, T.A.; Carrington, J.C. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 2010, 17, 997. [Google Scholar] [CrossRef]
- Chen, H.-M.; Chen, L.-T.; Patel, K.; Li, Y.-H.; Baulcombe, D.C.; Wu, S.-H. 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 2010, 107, 15269–15274. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Yang, G.S.; Chen, W.T.; Lin, R.M.; Ling, J.; Mao, Z.C.; Xie, B.Y. Characterization and function of Tomato yellow leaf curl virus-derived small RNAs generated in tolerant and susceptible tomato varieties. J. Integr. Agric. 2016, 15, 1785–1797. [Google Scholar] [CrossRef] [Green Version]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef]
- Addo-Quaye, C.; Eshoo, T.W.; Bartel, D.P.; Axtell, M.J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008, 18, 758–762. [Google Scholar] [CrossRef] [Green Version]
- Llave, C.; Xie, Z.; Kasschau, K.D.; Carrington, J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2002, 297, 2053–2056. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Billmeier, M.; Mohorianu, I.-I.; Green, D.; Fraser, W.; Dalmay, T. An improved protocol for small RNA library construction using high definition adapters. Methods Next Gener. Seq. 2015, 2, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Cloonan, N.; Wani, S.; Xu, Q.; Gu, J.; Lea, K.; Heater, S.; Barbacioru, C.; Steptoe, A.L.; Martin, H.C.; Nourbakhsh, E. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011, 12, R126. [Google Scholar] [CrossRef] [Green Version]
- Rotter, A.; Usadel, B.; Baebler, Š.; Stitt, M.; Gruden, K. Adaptation of the MapMan ontology to biotic stress responses: Application in solanaceous species. Plant Methods 2007, 3, 10. [Google Scholar] [CrossRef] [Green Version]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Hoorn, R.A.L.; Wulff, B.B.H.; Rivas, S.; Durrant, M.C.; van der Ploeg, A.; de Wit, P.J.G.M.; Jones, J.D.G. Structure–Function Analysis of Cf-9, a Receptor-Like Protein with Extracytoplasmic Leucine-Rich Repeats. Plant Cell 2005, 17, 1000–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuang, H.; Wei, F.; Marano, M.R.; Wirtz, U.; Wang, X.; Liu, J.; Shum, W.P.; Zaborsky, J.; Tallon, L.J.; Rensink, W.; et al. The R1 resistance gene cluster contains three groups of independently evolving, type I R1 homologues and shows substantial structural variation among haplotypes of Solanum demissum. Plant J. 2005, 44, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Fei, Z.; Joung, J.-G.; Tang, X.; Zheng, Y.; Huang, M.; Lee, J.M.; McQuinn, R.; Tieman, D.M.; Alba, R.; Klee, H.J.; et al. Tomato Functional Genomics Database: A comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res. 2011, 39, D1156–D1163. [Google Scholar] [CrossRef] [Green Version]
- Zuo, J.; Wang, Q.; Han, C.; Ju, Z.; Cao, D.; Zhu, B.; Luo, Y.; Gao, L. SRNAome and degradome sequencing analysis reveals specific regulation of sRNA in response to chilling injury in tomato fruit. Physiol. Plant 2017, 160, 142–154. [Google Scholar] [CrossRef]
- Pan, C.; Ye, L.; Zheng, Y.; Wang, Y.; Yang, D.; Liu, X.; Chen, L.; Zhang, Y.; Fei, Z.; Lu, G. Identification and expression profiling of microRNAs involved in the stigma exsertion under high-temperature stress in tomato. BMC Genomics 2017, 18, 843. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [Green Version]
- Chapman, E.J.; Prokhnevsky, A.I.; Gopinath, K.; Dolja, V.V.; Carrington, J.C. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 2004, 18, 1179–1186. [Google Scholar] [CrossRef] [Green Version]
- Lakatos, L.; Csorba, T.; Pantaleo, V.; Chapman, E.J.; Carrington, J.C.; Liu, Y.P.; Dolja, V.V.; Calvino, L.F.; López-Moya, J.J.; Burgyán, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006, 25, 2768–2780. [Google Scholar] [CrossRef] [Green Version]
- Shiboleth, Y.M.; Haronsky, E.; Leibman, D.; Arazi, T.; Wassenegger, M.; Whitham, S.A.; Gaba, V.; Gal-On, A. The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J. Virol. 2007, 81, 13135–13148. [Google Scholar] [CrossRef] [Green Version]
- Ye, K.; Malinina, L.; Patel, D.J. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 2003, 426, 874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chávez Montes, R.A.; Rosas-Cárdenas, D.F.F.; De Paoli, E.; Accerbi, M.; Rymarquis, L.A.; Mahalingam, G.; Marsch-Martínez, N.; Meyers, B.C.; Green, P.J.; de Folter, S. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat. Commun. 2014, 5, 3722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, D.-H. Functional diversity of microRNA variants in plants. J. Plant Biol. 2016, 59, 303–310. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, S.; Zhou, X.; Xia, J.; Chellappan, P.; Zhou, X.; Zhang, X.; Jin, H. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol. 2010, 11, R81. [Google Scholar] [CrossRef] [Green Version]
- Du, P.; Wu, J.; Zhang, J.; Zhao, S.; Zheng, H.; Gao, G.; Wei, L.; Li, Y. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 2011, 7, e1002176. [Google Scholar] [CrossRef]
- Hu, Q.; Hollunder, J.; Niehl, A.; Kørner, C.J.; Gereige, D.; Windels, D.; Arnold, A.; Kuiper, M.; Vazquez, F.; Pooggin, M.; et al. Specific Impact of Tobamovirus Infection on the Arabidopsis Small RNA Profile. PLoS ONE 2011, 6, e19549. [Google Scholar] [CrossRef] [Green Version]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Islam, W.; Qasim, M.; Noman, A.; Adnan, M.; Tayyab, M.; Farooq, T.H.; Wei, H.; Wang, L. Plant microRNAs: Front line players against invading pathogens. Microb. Pathog. 2018, 118, 9–17. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, F.; Li, J.; Chen, J.-P.; Zhang, H.-M. Integrative Analysis of the microRNAome and Transcriptome Illuminates the Response of Susceptible Rice Plants to Rice Stripe Virus. PLoS ONE 2016, 11, e0146946. [Google Scholar] [CrossRef] [Green Version]
- Shivaprasad, P.V.; Chen, H.-M.; Patel, K.; Bond, D.M.; Santos, B.A.; Baulcombe, D.C. A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef] [Green Version]
- Tsushima, D.; Adkar-Purushothama, C.R.; Taneda, A.; Sano, T. Changes in relative expression levels of viroid-specific small RNAs and microRNAs in tomato plants infected with severe and mild symptom-inducing isolates of Potato spindle tuber viroid. J. Gen. Plant Pathol. 2015, 81, 49–62. [Google Scholar] [CrossRef]
- Wang, W.; Luan, Y. The advance of tomato disease-related microRNAs. Plant Cell Rep. 2015, 34, 1089–1097. [Google Scholar] [CrossRef]
- Chiumenti, M.; Catacchio, C.R.; Miozzi, L.; Pirovano, W.; Ventura, M.; Pantaleo, V. A Short Indel-Lacking-Resistance Gene Triggers Silencing of the Photosynthetic Machinery Components Through TYLCSV-Associated Endogenous siRNAs in Tomato. Front. Plant Sci. 2018, 9, 1470. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Jeong, D.-H.; De Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; González, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Wang, J.; Tung, J.; Liu, D.; Zhou, Y.; He, S.; Du, Y.; Baker, B.; Li, F. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018, 14, e1006756. [Google Scholar] [CrossRef]
- Seo, J.K.; Wu, J.; Lii, Y.; Li, Y.; Jin, H. Contribution of small RNA pathway components in plant immunity. Mol. Plant Microbe Interact. 2013, 26, 617–625. [Google Scholar] [CrossRef]
- Park, J.H.; Shin, C. The role of plant small RNAs in NB-LRR regulation. Brief. Funct. Genomics 2015, 14, 268–274. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, V.M.; Muller, S.; Baulcombe, D.; Puigdomenech, P. Evolution of NBS-LRR gene copies among Dicot plants and its regulation by members of the miR482/2118 superfamily of miRNAs. Mol. Plant 2015, 8, 329–331. [Google Scholar] [CrossRef] [Green Version]
- Canto-Pastor, A.; Santos, B.A.; Valli, A.A.; Summers, W.; Schornack, S.; Baulcombe, D.C. Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc. Natl. Acad. Sci. USA 2019, 116, 2755–2760. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Hardcastle, T.J.; Canto Pastor, A.; Yip, W.H.; Tang, S.; Baulcombe, D.C. A novel DCL2-dependent miRNA pathway in tomato affects susceptibility to RNA viruses. Genes Dev. 2018, 32, 1155–1160. [Google Scholar] [CrossRef] [Green Version]
- Bengyella, L.; Waikhom, S.D.; Allie, F.; Rey, C. Virus tolerance and recovery from viral induced-symptoms in plants are associated with transcriptome reprograming. Plant Mol. Biol. 2015, 89, 243–252. [Google Scholar] [CrossRef] [PubMed]





| Mk-21 1 | PVY-21 1 | Mk-30 1 | PVY-30 1 | |||||
| Redundant Reads Number | Nonredundant Reads Number | Redundant Reads Number | Nonredundant Reads Number | Redundant Reads Number | Nonredundant Reads Number | Redundant Reads Number | Nonredundant Reads Number | |
| raw reads (library size) | 28.222.339 | 6.959.896 | 23.645.346 | 3.744.089 | 17.602.784 | 4.531.582 | 12.801.610 | 3.164.947 |
| low-complexity sequences | 195 | 171 | 105 | 93 | 172 | 148 | 134 | 118 |
| clean reads 18–26 nt | 22.463.045 | 6.376.853 | 21.507.027 | 3.392.422 | 13.973.890 | 4.137.134 | 10.945.324 | 2.845.983 |
| Length Distribution of Clean Reads | Redundant Reads Number | % 2 | Redundant Reads Number | % 2 | Redundant Reads Number | % 2 | Redundant Reads Number | % 2 |
| 18 | 207.038 | 0.92 | 201.139 | 0.94 | 151.081 | 1.08 | 137.259 | 1.25 |
| 19 | 280.484 | 1.25 | 288.476 | 1.34 | 175.548 | 1.26 | 182.771 | 1.67 |
| 20 | 495.843 | 2.21 | 484.206 | 2.25 | 300.467 | 2.15 | 281.912 | 2.58 |
| 21 | 2.133.848 | 9.5 | 12.018.765 | 55.88 | 1.661.996 | 11.89 | 4.418.744 | 40.37 |
| 22 | 2.226.996 | 9.91 | 3.473.057 | 16.15 | 1.622.666 | 11.61 | 1.679.439 | 15.34 |
| 23 | 3.475.721 | 15.47 | 1.266.933 | 5.89 | 2.060.721 | 14.75 | 1.018.064 | 9.3 |
| 24 | 12.562.212 | 55.92 | 3.488.376 | 16.22 | 7.372.172 | 52.76 | 2.930.233 | 26.77 |
| 25 | 761.854 | 3.39 | 197.821 | 0.92 | 425.439 | 3.04 | 198.083 | 1.81 |
| 26 | 319.049 | 1.42 | 88.254 | 0.41 | 203.800 | 1.46 | 98.819 | 0.9 |
| tomato SL3.0 genome mapped reads (100% identity) | 20.289.212 | 90.32 | 10.134.178 | 47.12 | 12.632.820 | 90.40 | 6.506.699 | 59.45 |
| noncoding RNAs | 1787544 | 7.96 | 558036 | 2.59 | 1157563 | 8.28 | 554155 | 5.06 |
| rRNAs | 1653355 | 7.36 | 519130 | 2.41 | 1034369 | 7.40 | 516761 | 4.72 |
| tRNAs | 73891 | 0.33 | 25743 | 0.12 | 95440 | 0.68 | 22052 | 0.20 |
| snoRNA | 48703 | 0.22 | 6736 | 0.03 | 20753 | 0.15 | 10516 | 0.10 |
| snRNA | 6507 | 0.03 | 3602 | 0.02 | 4221 | 0.03 | 2674 | 0.02 |
| lncRNA | 5088 | 0.02 | 2825 | 0.01 | 2780 | 0.02 | 2152 | 0.02 |
| Tot. | 1787544 | 7.96 | 558036 | 2.59 | 1157563 | 8.28 | 554155 | 5.06 |
| tomato phasiRNAs all | 53902 | 0.27 | 50163 | 0.49 | 20991 | 0.17 | 19981 | 0.31 |
| tomato phasiRNAs PVY specific | 0 | 0 | 313 | 0.003 | 0 | 0 | 219 | 0.003 |
| known mature miRNAs | 758288 | 3.38 | 883482 | 4.11 | 810148 | 5.80 | 455002 | 4.16 |
| vsiRNAs (100% identity) | 928 | 0.004 | 10040027 | 46.68 | 590 | 0.004 | 3593569 | 32.83 |
| sequence length of vsiRNAs | abs | % 3 | abs | % 3 | abs | % 3 | abs | % 3 |
| 18 | 2 | 0.22 | 17649 | 0.18 | 7299 | 0.20 | ||
| 19 | 5 | 0.54 | 24487 | 0.24 | 1 | 0.17 | 9812 | 0.27 |
| 20 | 9 | 0.97 | 66285 | 0.66 | 5 | 0.85 | 24028 | 0.67 |
| 21 | 726 | 78.23 | 8209564 | 81.77 | 469 | 79.49 | 2914094 | 81.09 |
| 22 | 179 | 19.29 | 1652385 | 16.46 | 109 | 18.47 | 616990 | 17.17 |
| 23 | 4 | 0.43 | 41968 | 0.42 | 4 | 0.68 | 13838 | 0.39 |
| 24 | 3 | 0.32 | 24511 | 0.24 | 2 | 0.34 | 6138 | 0.17 |
| 25 | 2405 | 0.02 | 984 | 0.03 | ||||
| 26 | 773 | 0.01 | 386 | 0.01 | ||||
| Normalized Read Counts (RPM) | Relative Accumulation (PVY vs. Mk) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | Length | miRNA ID | Mk-21 | PVY-21 | Mk-30 | PVY-30 | Log2fc 21 dpi | Log2fc 30 dpi | PVY-Associated 2 |
| 1 | 22 | sly-miR6023 | 257.9 | 161.2 | 118.1 | 177.5 | −0.7 | 0.6 | |
| 1 | 23 | sly-miR6023.30 | 4.3 | 154.3 | 1.3 | 185.2 | 5.2 | 7.2 | |
| 1 | 19 | sly-miR6023.3 | 0.2 | 46.1 | 0.1 | 31.0 | 7.9 | 9.0 | |
| 1 | 23 | sly-miR6023.14 | 0.0 | 4.1 | 0.1 | 4.5 | 6.8 | 5.2 | YES |
| 1 | 23 | sly-miR6023.16 | 0.1 | 2.9 | 0.1 | 5.6 | 4.3 | 6.5 | YES |
| 1 | 22 | sly-miR6023.7 | 0.0 | 2.6 | 0.0 | 2.5 | 7.1 | 6.4 | YES |
| 1 | 22 | sly-miR6023.13 | 0.1 | 1.7 | 0.0 | 1.1 | 4.5 | 5.2 | YES |
| 1 | 22 | sly-miR6023.21 | 0.0 | 1.7 | 0.0 | 2.5 | 6.5 | 6.4 | YES |
| 1 | 21 | sly-miR6023.29 | 0.7 | 0.5 | 0.4 | 0.6 | |||
| 2 | 22 | sly-miR6023.1 | 0.6 | 59.3 | 0.6 | 51.0 | 6.6 | 6.4 | |
| 2 | 21 | sly-miR6023.4 | 0.2 | 1.4 | 0.2 | 1.1 | 2.9 | 2.7 | |
| 2 | 24 | sly-miR6023.5 | 3.4 | 0.6 | 2.3 | 1.3 | −2.4 | −0.8 | |
| 3 | 22 | sly-miR6023.28 | 13.5 | 87.2 | 13.9 | 96.9 | 2.7 | 2.8 | |
| 3 | 21 | sly-miR6023.18 | 0.7 | 15.3 | 1.5 | 12.0 | 4.4 | 3.0 | |
| 3 | 21 | sly-miR6023.17 | 0.1 | 14.8 | 0.0 | 9.1 | 7.6 | 8.2 | YES |
| 3 | 22 | sly-miR6023.10 | 0.1 | 9.0 | 0.0 | 12.2 | 6.3 | 8.7 | YES |
| 3 | 21 | sly-miR6023.19 | 0.4 | 6.3 | 0.8 | 5.2 | 3.8 | 2.7 | |
| 3 | 21 | sly-miR6023.27 | 0.6 | 5.8 | 0.5 | 8.9 | 3.4 | 4.1 | |
| 3 | 21 | sly-miR6023.25 | 0.1 | 5.3 | 0.1 | 6.1 | 5.6 | 5.7 | YES |
| 3 | 21 | sly-miR6023.2 | 0.3 | 3.8 | 0.5 | 3.6 | 3.7 | 2.9 | |
| 3 | 22 | sly-miR6023.26 | 0.0 | 3.6 | 0.0 | 6.0 | 7.6 | 7.7 | YES |
| 3 | 23 | sly-miR6023.11 | 0.0 | 1.3 | 0.3 | 1.3 | 5.1 | 2.1 | YES |
| 4 | 21 | sly-miR6023.15 | 0.1 | 2.2 | 0.1 | 4.2 | 4.9 | 5.1 | YES |
| 4 | 21 | sly-miR6023.12 | 0.0 | 1.4 | 0.0 | 1.1 | 6.2 | 5.1 | YES |
| 5 | 19 | sly-miR6023.23 | 1.6 | 43.1 | 2.6 | 47.7 | 4.7 | 4.2 | |
| 5 | 20 | sly-miR6023.24 | 0.1 | 5.9 | 0.0 | 7.1 | 5.3 | 7.9 | YES |
| 5 | 20 | sly-miR6023.8 | 0.1 | 5.4 | 0.1 | 6.3 | 5.6 | 5.7 | YES |
| 5 | 18 | sly-miR6023.22 | 0.1 | 2.4 | 0.1 | 2.0 | 4.0 | 5.1 | YES |
| 5 | 21 | sly-miR6023.20 | 0.3 | 2.0 | 1.1 | 1.5 | 2.8 | 0.4 | |
| 5 | 22 | sly-miR6023.6 | 0.1 | 1.3 | 0.0 | 1.5 | 3.6 | 5.7 | YES |
| 5 | 21 | sly-miR6023.9 | 0.0 | 1.3 | 0.1 | 2.3 | 6.2 | 5.2 | YES |
| Normalized Read Counts (RPM) | Relative Accumulation (PVY vs. Mk) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sequence | miRNA ID 1 | Identical miRNA ID in miRBase 2 | miRNA Family | Mk-21 | PVY-21 | Mk-30 | PVY-30 | Log2fc 21 dpi | Log2fc 30 dpi |
| GGAGGCAGCGGTTCATCGATC | novel_sly-miR162a* | aly-miR162a-5p | miR162 | 6.2 | 130.8 | 3.2 | 96.9 | 4.4 | 4.9 |
| AGATCATGTGGTTGCTTCACC | novel_sly-miR167d | NO | miR167 | 1.0 | 36.0 | 0.5 | 30.1 | 5.1 | 5.8 |
| TTCCAAAGCTGCAGAAATGAGT | novel_sly-miR8033 | stu-miR8033-5p | miR8033 | 1.2 | 17.8 | 2.8 | 12.7 | 3.9 | 2.2 |
| TTTTGTAGTAACTGTACCACA | novel-sly-miR1 | NO | novel | 0.1 | 2.9 | 0.0 | 2.3 | 4.3 | 6.2 |
| GGGGCAACTTGAGATCACATG | novel-sly-miR11 | NO | novel | 295.5 | 19,873 | 184.5 | 4511.7 | 6.1 | 4.6 |
| TGTGTTCTCAGGTTACCCCTG | novel-sly-miR11* | NO | novel | 254.1 | 9302.3 | 187.6 | 4058.3 | 5.2 | 4.4 |
| AACTGTCGGGAGACATTAGCT | novel-sly-miR2 | NO | novel | 1.1 | 8.4 | 1.6 | 7.1 | 2.9 | 2.2 |
| TACTATCTGATTTAAGATTAG | novel-sly-miR3 | NO | novel | 0.0 | 4.5 | 0.0 | 3.6 | 7.9 | 6.9 |
| CCTGAACTATCACCATCTATG | novel-sly-miR5 | NO | novel | 0.1 | 7.8 | 0.0 | 5.3 | 6.1 | 7.5 |
| TGGCAAGTAAGCGCTCCAACT | novel-sly-miR6 | NO | novel | 0.0 | 4.3 | 0.2 | 3.8 | 6.8 | 4.4 |
| AGGGGAGATAGATGAAGTTAGG | novel-sly-miR7 | NO | novel | 0.2 | 7.3 | 0.2 | 7.1 | 5.0 | 5.3 |
| TTATTTGGGGTAGATGAGCTC | novel-sly-miR8 | NO | novel | 1.0 | 14.3 | 0.4 | 9.5 | 3.9 | 4.7 |
| AACAACTGATAGTTGAGGTGT | novel-sly-miR9 | NO | novel | 0.3 | 7.5 | 0.0 | 4.8 | 4.8 | 7.3 |
| ATTTACCCCAAGTTCGTTGTC | sly-miR10537 | sly-miR10537 | miR10537 | 1.2 | 14.1 | 0.9 | 8.4 | 3.6 | 3.2 |
| ATAATAACTATTAGTTGAATG | sly-miR10540 | sly-miR10540 | miR10540 | 0.0 | 1.5 | 0.0 | 1.2 | 6.3 | 5.3 |
| CATGTGCCTGTTTTCCCCATC | sly-miR164a-3p | sly-miR164a-3p | miR164 | 0.9 | 30.8 | 1.0 | 14.9 | 5.1 | 4.0 |
| GGGATGTTGTCTGGCTCGACA | sly-miR166c-5p | sly-miR166c-5p | miR166 | 155.7 | 1440.6 | 99.9 | 905.8 | 3.2 | 3.2 |
| AGGTCATCTAGCAGCTTCAAT | sly-miR167b-3p | sly-miR167b-3p | miR167 | 0.2 | 9.5 | 0.1 | 7.0 | 5.4 | 6.9 |
| CCTGCCTTGCATCAACTGAAT | sly-miR168a-3p | sly-miR168a-3p | miR168 | 67.3 | 441.4 | 32.9 | 201.2 | 2.7 | 2.6 |
| CCCGCCTTGCATCAACTGAAT | sly-miR168b-3p | sly-miR168b-3p | miR168 | 220.1 | 1731.6 | 121.5 | 794.4 | 3.0 | 2.7 |
| TTGAGCCGTGCCAATATCACG | sly-miR171b-3p | sly-miR171b-3p | miR171 | 3.4 | 28.0 | 1.4 | 22.0 | 3.0 | 4.0 |
| TATTGGCCTGGTTCACTCAGA | sly-miR171f | sly-miR171f | miR171 | 36.6 | 371.0 | 25.1 | 226.4 | 3.3 | 3.2 |
| ACGAGAGTCATCTGTGACAGG | sly-miR1919a|sly-miR1919c-3p|sly-miR1919b|novel_sly-miR1919d | sly-miR1919a | miR1919 | 9.0 | 93.2 | 9.0 | 52.8 | 3.4 | 2.6 |
| AGGAAACTGTTTAGTCCAACC | sly-miR319d | sly-miR319d | miR319 | 0.0 | 1.2 | 0.0 | 1.6 | 5.0 | 5.8 |
| CGCTATCCATCCTGAGTTTTA | sly-miR390a-3p | sly-miR390a-3p | miR390 | 0.5 | 3.9 | 0.2 | 6.1 | 3.0 | 4.7 |
| AAGCTCAGGAGGGATAGCACC | sly-miR390a-5p | sly-miR390a-5p | miR390 | 75.9 | 524.5 | 70.9 | 378.0 | 2.8 | 2.4 |
| CGCTATCCATCCTGAGTTTCA | sly-miR390b-3p | sly-miR390b-3p | miR390 | 0.1 | 1.6 | 0.0 | 1.1 | 4.4 | 5.2 |
| ATCATGCGATCTCTTCGGAAT | sly-miR393 | sly-miR393 | miR393 | 10.0 | 120.2 | 6.7 | 76.1 | 3.6 | 3.5 |
| AGGTGGGCATACTGTCAACA | sly-miR394-3p | sly-miR394-3p | miR394 | 1.3 | 16.4 | 1.6 | 28.5 | 3.7 | 4.2 |
| GTTCAATAAAGCTGTGGGAAG | sly-miR396a-3p | sly-miR396a-3p | miR396 | 49.9 | 1727.2 | 45.5 | 1132.1 | 5.1 | 4.6 |
| ATTGAGTGCAGCGTTGATGA | sly-miR397-5p | sly-miR397-5p | miR397 | 10.0 | 359.3 | 15.5 | 208.7 | 5.2 | 3.7 |
| TATGTTCTCAGGTCGCCCCTG | sly-miR398a | sly-miR398a | miR398 | 834.8 | 13869 | 925.1 | 5806.1 | 4.1 | 2.6 |
| CGTTTGTGCGTGAATCTAACA | sly-miR403-5p | sly-miR403-5p | miR403 | 0.7 | 11.9 | 0.2 | 9.7 | 4.1 | 5.3 |
| ACGGGGACGAGCCAGAGCATG | sly-miR408 | sly-miR408 | miR408 | 1.6 | 14.5 | 1.0 | 40.2 | 3.2 | 5.4 |
| TGTGGGTGGGGTGGAAAGATT | sly-miR482e-5p | sly-miR482e-5p | miR482 | 6.0 | 89.3 | 9.6 | 202.5 | 3.9 | 4.4 |
| AGGTGTAGGTGTTCATGCAGA | sly-miR530 | sly-miR530 | miR530 | 0.4 | 48.7 | 0.4 | 18.7 | 6.8 | 5.7 |
| ATGGGTAGCACAAGGATTAATG | sly-miR6027-5p | sly-miR6027-5p | miR6027 | 313.9 | 4006.6 | 281.9 | 3442.1 | 3.7 | 3.6 |
| TGAAATCCATGAGCCTAAACT | sly-miR9470-5p | sly-miR9470-5p | miR9470 | 0.7 | 13.6 | 0.5 | 5.3 | 4.2 | 3.5 |
| TTTCAGTAGACGTTGTGAATA | sly-miR9472-5p | sly-miR9472-5p | miR9472 | 0.2 | 4.3 | 0.2 | 2.9 | 4.5 | 4.0 |
| TGTAGAAGTCATGAATAAAATG | sly-miR9474-5p | sly-miR9474-5p | miR9474 | 6.3 | 26.5 | 6.0 | 33.6 | 2.1 | 2.5 |
| AAAAAGATGCAGGACTAGACC | sly-miR9476-3p | sly-miR9476-3p | miR9476 | 114.0 | 675.9 | 112.5 | 493.2 | 2.6 | 2.1 |
| AACAACATACTTACTGAAATGCCA | novel_sly-miR8020 | NO | miR8020 | 6.6 | 1.3 | 5.5 | 2.7 | −2.3 | −1.0 |
| GAATTTCATTGAGTATGTTGTTGT | novel_sly-miR8020* | NO | miR8020 | 1.6 | 0.3 | 0.6 | 0.1 | −2.6 | − |
| AGTGGACAAGTAAAGGTGGATGGA | novel-sly-miR4 | NO | novel | 5.2 | 0.9 | 3.3 | 1.1 | −2.6 | −1.5 |
| AACGAGTGAGACTTGCTCAGTTGG | sly-miR10529 | sly-miR10529 | miR10529 | 1.6 | 0.3 | 0.4 | 0.8 | −2.6 | − |
| ACGTCCCTTCCCCATCGTTCAACA | sly-miR10530 | sly-miR10530 | miR10530 | 3.7 | 0.7 | 2.7 | 1.3 | −2.3 | −1.1 |
| TTTTAGCAAGAGTTGTTTTACC | sly-miR6024 | sly-miR6024 | miR6024 | 365.2 | 56.1 | 158.0 | 90.1 | −2.7 | −0.8 |
| AAGTGTGTCTCTGGAATTTCGGGC | sly-miR7981f | sly-miR7981f | miR7981 | 5.2 | 1.1 | 3.4 | 2.5 | −2.2 | −0.4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prigigallo, M.I.; Križnik, M.; De Paola, D.; Catalano, D.; Gruden, K.; Finetti-Sialer, M.M.; Cillo, F. Potato Virus Y Infection Alters Small RNA Metabolism and Immune Response in Tomato. Viruses 2019, 11, 1100. https://doi.org/10.3390/v11121100
Prigigallo MI, Križnik M, De Paola D, Catalano D, Gruden K, Finetti-Sialer MM, Cillo F. Potato Virus Y Infection Alters Small RNA Metabolism and Immune Response in Tomato. Viruses. 2019; 11(12):1100. https://doi.org/10.3390/v11121100
Chicago/Turabian StylePrigigallo, Maria I., Maja Križnik, Domenico De Paola, Domenico Catalano, Kristina Gruden, Mariella M. Finetti-Sialer, and Fabrizio Cillo. 2019. "Potato Virus Y Infection Alters Small RNA Metabolism and Immune Response in Tomato" Viruses 11, no. 12: 1100. https://doi.org/10.3390/v11121100