Bacteriophage Sf6 Tailspike Protein for Detection of Shigella flexneri Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Bacterial Strains
2.2. Cloning and Protein Purification
2.3. Oligo- and Polysaccharide Preparation
2.4. ELISA Like Tailspike Adsorption (ELITA) Assay
2.5. Fluorescence Labeling
2.6. Fluorescence Spectroscopy
2.7. Surface Plasmon Resonance
3. Results
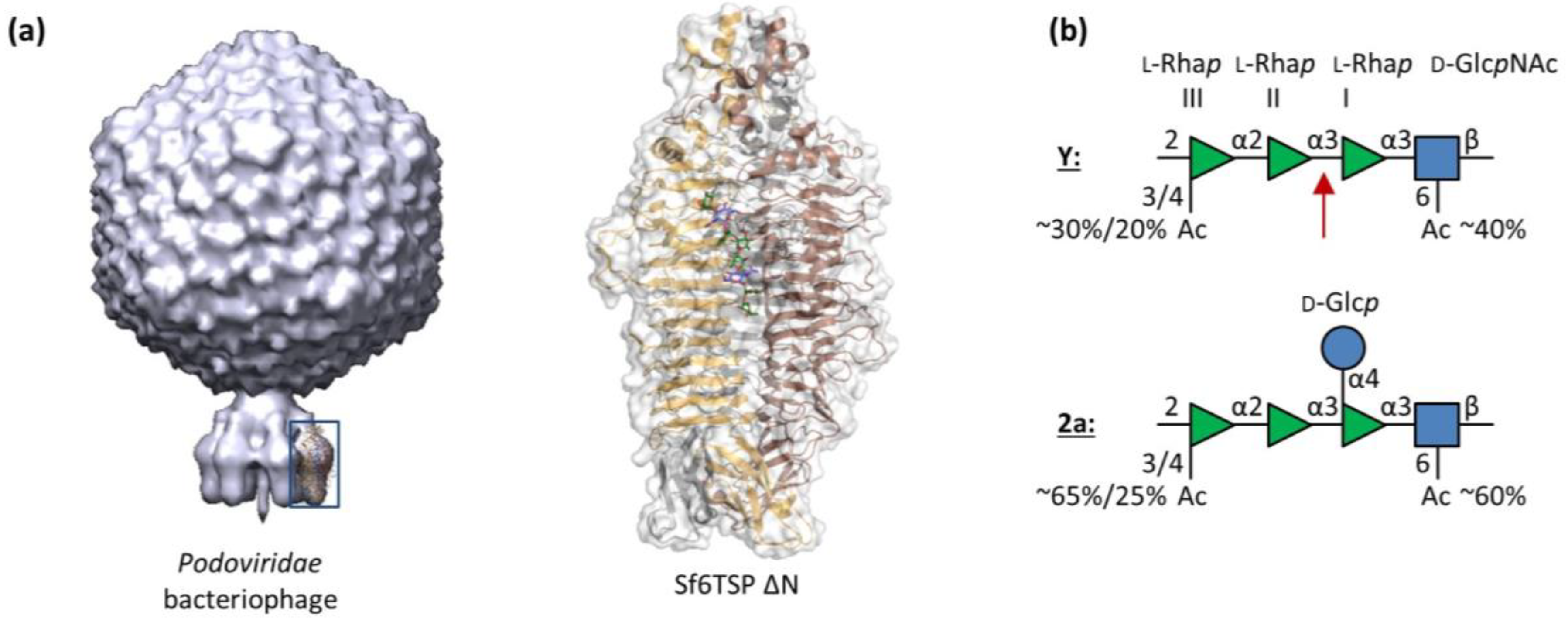
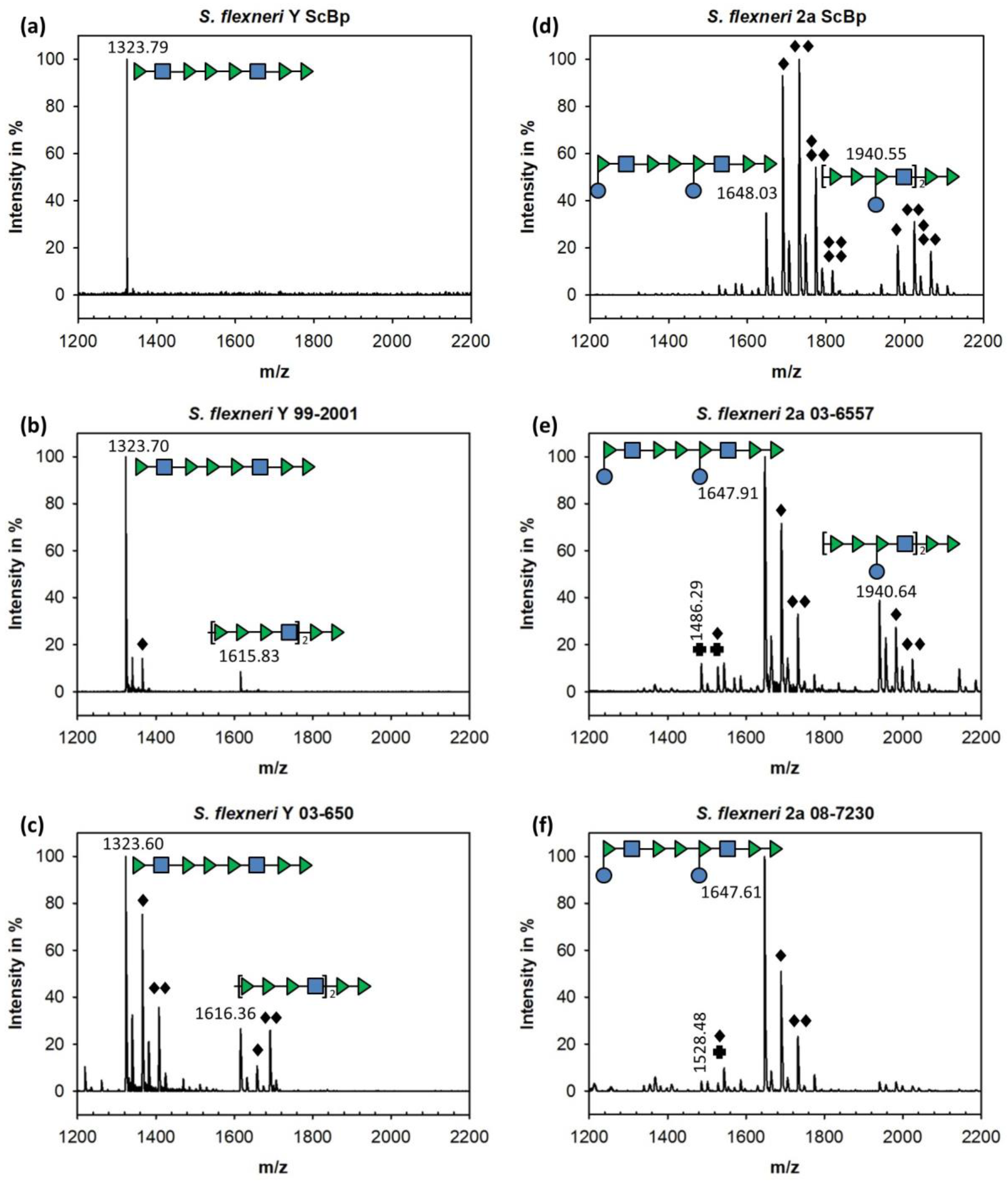
3.1. Sf6TSP Binds and Enzymatically Cleaves the S. flexneri Serotype 2a O-Antigen
3.2. Binding of Sf6TSP to S. flexneri O-Antigens with O-Acetyl and Glucosyl Modifications
3.3. ELITA: ELISA Like Tailspike Adsorption Assay for Detection of S. flexneri
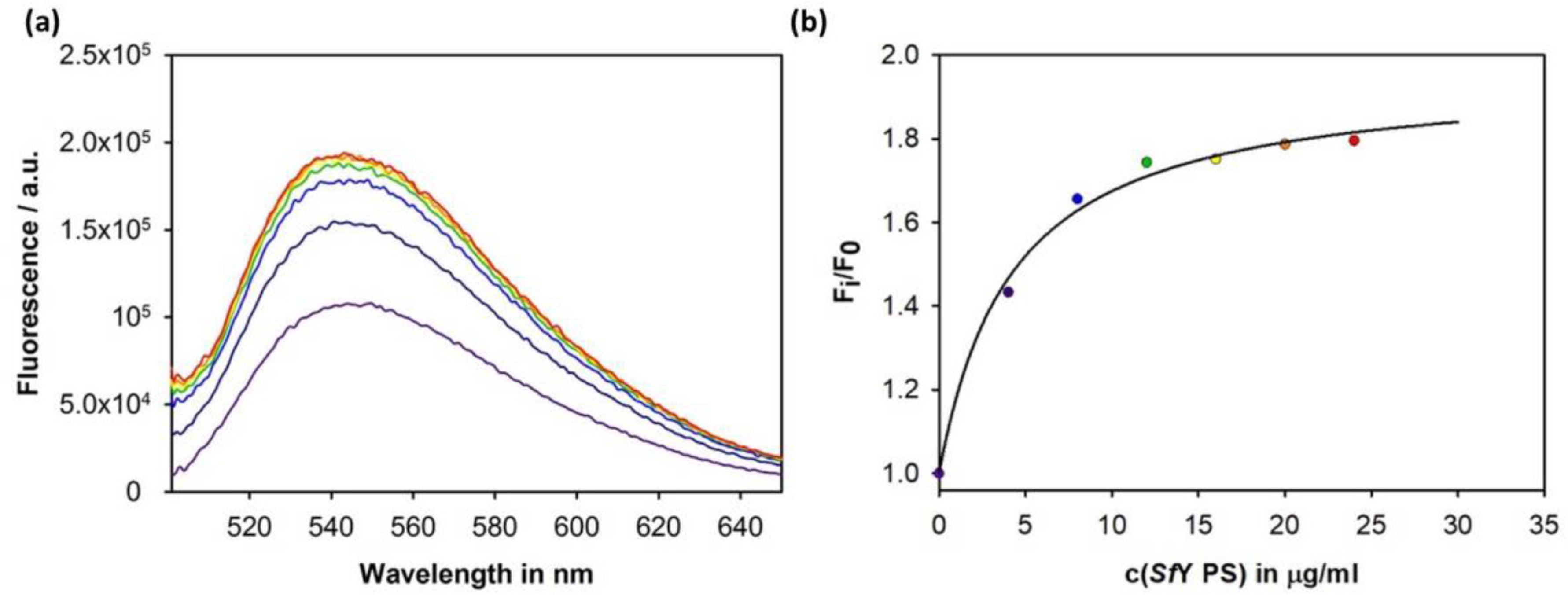
3.4. Construction of an Sf6TSP Probe with O-Antigen Specific Fluorescence Amplitude Increase
4. Discussion
4.1. Binding Specificity of the Sf6TSP Endorhamnosidase
4.2. Sf6TSP as a Pathogen Sensor for S. flexneri
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pires, S.M.; Fischer-Walker, C.L.; Lanata, C.F.; Devleesschauwer, B.; Hall, A.J.; Kirk, M.D.; Duarte, A.S.R.; Black, R.E.; Angulo, F.J. Aetiology-Specific Estimates of the Global and Regional Incidence and Mortality of Diarrhoeal Diseases Commonly Transmitted through Food. PLoS ONE 2015, 10, e0142927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef]
- Sethuvel, D.P.M.; Ragupathi, N.K.D.; Anandan, S.; Veeraraghavan, B. Update on: Shigella new serogroups/serotypes and their antimicrobial resistance. Lett. Appl. Microbiol. 2017, 64, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Hale, T.L.; Keusch, G.T. Shigella. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; ISBN 978-0-9631172-1-2. [Google Scholar]
- Penatti, M.P.A.; Hollanda, L.M.; Nakazato, G.; Campos, T.A.; Lancellotti, M.; Angellini, M.; Brocchi, M.; Rocha, M.M.M.; Silveira, W.D. Epidemiological characterization of resistance and PCR typing of Shigella flexneri and Shigella sonnei strains isolated from bacillary dysentery cases in Southeast Brazil. Braz. J. Med. Biol. Res. 2007, 40, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Roberts, A.P.; Xu, X.; Yang, C.; Yang, X.; Wang, J.; Yi, S.; Li, Y.; Ma, Q.; Wu, F.; et al. Transferable Plasmid-Borne MCR-1 in a Colistin-Resistant Shigella flexneri Isolate. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Lan, R.; Alles, M.C.; Donohoe, K.; Martinez, M.B.; Reeves, P.R. Molecular Evolutionary Relationships of Enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 2004, 72, 5080–5088. [Google Scholar] [CrossRef] [PubMed]
- Bliven, K.A.; Maurelli, A.T. Evolution of Bacterial Pathogens within the Human Host. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Van den Beld, M.J.C.; Reubsaet, F.A.G. Differentiation between Shigella, enteroinvasive Escherichia coli (EIEC) and noninvasive Escherichia coli. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Health Canada. The Compendium of Analytical Methods. Available online: https://www.canada.ca/en/health-canada/services/food-nutrition/research-programs-analytical-methods/analytical-methods/compendium-methods.html (accessed on 15 March 2018).
- DuPont, H.L.; Levine, M.M.; Hornick, R.B.; Formal, S.B. Inoculum size in shigellosis and implications for expected mode of transmission. J. Infect. Dis. 1989, 159, 1126–1128. [Google Scholar] [CrossRef] [PubMed]
- Duran, C.; Nato, F.; Dartevelle, S.; Thi Phuong, L.N.; Taneja, N.; Ungeheuer, M.N.; Soza, G.; Anderson, L.; Benadof, D.; Zamorano, A.; et al. Rapid Diagnosis of Diarrhea Caused by Shigella sonnei Using Dipsticks; Comparison of Rectal Swabs, Direct Stool and Stool Culture. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Shabarova, T.; Eugster, M.R.; Eichenseher, F.; Tchang, V.S.; Banz, M.; Loessner, M.J. Rapid Multiplex Detection and Differentiation of Listeria Cells by Use of Fluorescent Phage Endolysin Cell Wall Binding Domains. Appl. Environ. Microbiol. 2010, 76, 5745–5756. [Google Scholar] [CrossRef] [PubMed]
- Fujinami, Y.; Hirai, Y.; Sakai, I.; Yoshino, M.; Yasuda, J. Sensitive Detection of Bacillus anthracis Using a Binding Protein Originating from γ-Phage. Microbiol. Immunol. 2007, 51, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Schofield, D.A.; Wray, D.J.; Molineux, I.J. Isolation and development of bioluminescent reporter phages for bacterial dysentery. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 34, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Yim, P.B.; Clarke, M.L.; McKinstry, M.; Lacerda, S.H.D.P.; Pease, L.F.; Dobrovolskaia, M.A.; Kang, H.; Read, T.D.; Sozhamannan, S.; Hwang, J. Quantitative characterization of quantum dot-labeled λ phage for Escherichia coli detection. Biotechnol. Bioeng. 2009, 104, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Peltomaa, R.; López-Perolio, I.; Benito-Peña, E.; Barderas, R.; Moreno-Bondi, M.C. Application of bacteriophages in sensor development. Anal. Bioanal. Chem. 2016, 408, 1805–1828. [Google Scholar] [CrossRef] [PubMed]
- Doore, S.M.; Schrad, J.R.; Dean, W.F.; Dover, J.A.; Parent, K.N. Shigella Phages Isolated during a Dysentery Outbreak Reveal Uncommon Structures and Broad Species Diversity. J. Virol. 2018, 92, e02117-17. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.R.; Molineux, I.J. Short Noncontractile Tail Machines: Adsorption and DNA Delivery by Podoviruses. In Viral Molecular Machines; Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 2012; pp. 143–179. ISBN 978-1-4614-0979-3. [Google Scholar]
- Leiman, P.G.; Shneider, M.M. Contractile Tail Machines of Bacteriophages. In Viral Molecular Machines; Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 2012; pp. 93–114. ISBN 978-1-4614-0979-3. [Google Scholar]
- Davidson, A.R.; Cardarelli, L.; Pell, L.G.; Radford, D.R.; Maxwell, K.L. Long Noncontractile Tail Machines of Bacteriophages. In Viral Molecular Machines; Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 2012; pp. 115–142. ISBN 978-1-4614-0979-3. [Google Scholar]
- Broeker, N.K.; Barbirz, S. Not a barrier but a key: How bacteriophages exploit host’s O-antigen as an essential receptor to initiate infection. Mol. Microbiol. 2017, 105, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Broeker, N.K.; Andres, D.; Kang, Y.; Gohlke, U.; Schmidt, A.; Kunstmann, S.; Santer, M.; Barbirz, S. Complex carbohydrate recognition by proteins: Fundamental insights from bacteriophage cell adhesion systems. Perspect. Sci. 2017, 11, 45–52. [Google Scholar] [CrossRef]
- Schmidt, A.; Rabsch, W.; Broeker, N.K.; Barbirz, S. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC Microbiol. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Barbirz, S.; Becker, M.; Freiberg, A.; Seckler, R. Phage Tailspike Proteins with β-Solenoid Fold as Thermostable Carbohydrate Binding Materials. Macromol. Biosci. 2009, 9, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [PubMed]
- Binns, M.M.; Vaughan, S.; Timmis, K.N. O-antigens are essential virulence factors of Shigella sonnei and Shigella dysenteriae 1. Zentralbl. Bakteriol. Mikrobiol. Hyg. B 1985, 181, 197–205. [Google Scholar] [PubMed]
- West, N.P.; Sansonetti, P.; Mounier, J.; Exley, R.M.; Parsot, C.; Guadagnini, S.; Prévost, M.-C.; Prochnicka-Chalufour, A.; Delepierre, M.; Tanguy, M.; et al. Optimization of Virulence Functions Through Glucosylation of Shigella LPS. Science 2005, 307, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Freiberg, A.; Morona, R.; Van Den Bosch, L.; Jung, C.; Behlke, J.; Carlin, N.; Seckler, R.; Baxa, U. The Tailspike Protein of Shigella Phage Sf6: A Structural Homolog of Salmonella Phage P22 Tailspike Protein without Sequence Similarity in the β-helix Domain. J. Biol. Chem. 2003, 278, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Gohlke, U.; Engström, O.; Hamark, C.; Scheidt, T.; Kunstmann, S.; Heinemann, U.; Widmalm, G.; Santer, M.; Barbirz, S. Bacteriophage Tailspikes and Bacterial O-Antigens as a Model System to Study Weak-Affinity Protein–Polysaccharide Interactions. J. Am. Chem. Soc. 2016, 138, 9109–9118. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.J.; Barbirz, S.; Heinle, K.; Freiberg, A.; Seckler, R.; Heinemann, U. An Intersubunit Active Site between Supercoiled Parallel β Helices in the Trimeric Tailspike Endorhamnosidase of Shigella flexneri Phage Sf6. Structure 2008, 16, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Gemski, P.; Koeltzow, D.E.; Formal, S.B. Phage conversion of Shigella flexneri group antigens. Infect. Immun. 1975, 11, 685–691. [Google Scholar] [PubMed]
- Lindberg, A.A.; Wollin, R.; Gemski, P.; Wohlhieter, J.A. Interaction between bacteriophage Sf6 and Shigella Flexneri. J. Virol. 1978, 27, 38–44. [Google Scholar] [PubMed]
- Casjens, S.; Winn-Stapley, D.A.; Gilcrease, E.B.; Morona, R.; Kühlewein, C.; Chua, J.E.H.; Manning, P.A.; Inwood, W.; Clark, A.J. The Chromosome of Shigella flexneri Bacteriophage Sf6: Complete Nucleotide Sequence, Genetic Mosaicism, and DNA Packaging. J. Mol. Biol. 2004, 339, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Chua, J.E.; Manning, P.A.; Morona, R. The Shigella flexneri bacteriophage Sf6 tailspike protein (TSP)/endorhamnosidase is related to the bacteriophage P22 TSP and has a motif common to exo- and endoglycanases, and C-5 epimerases. Microbiol. Read. Engl. 1999, 145 Pt 7, 1649–1659. [Google Scholar] [CrossRef]
- Perepelov, A.V.; Shekht, M.E.; Liu, B.; Shevelev, S.D.; Ledov, V.A.; Senchenkova, S.N.; L’vov, V.L.; Shashkov, A.S.; Feng, L.; Aparin, P.G.; et al. Shigella flexneri O-antigens revisited: Final elucidation of the O-acetylation profiles and a survey of the O-antigen structure diversity. FEMS Immunol. Med. Microbiol. 2012, 66, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Knirel, Y.A.; Lan, R.; Senchenkova, S.N.; Luo, X.; Perepelov, A.V.; Wang, Y.; Shashkov, A.S.; Xu, J.; Sun, Q. Identification of an O-Acyltransferase Gene (OACB) That Mediates 3- and 4-O-Acetylation of Rhamnose III in Shigella flexneri O Antigens. J. Bacteriol. 2014, 196, 1525–1531. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, C.; Chassagne, P.; Theillet, F.-X.; Guerreiro, C.; Thouron, F.; Nato, F.; Delepierre, M.; Sansonetti, P.J.; Phalipon, A.; Mulard, L.A. Non-stoichiometric O-acetylation of Shigella flexneri 2a O-specific polysaccharide: Synthesis and antigenicity. Org. Biomol. Chem. 2014, 12, 4218–4232. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.A.; Beltrame, J.; Manning, P.A. The OAC gene encoding a lipopolysaccharide O-antigen acetylase maps adjacent to the integrase-encoding gene on the genome of Shigella flexneri bacteriophage Sf6. Gene 1991, 107, 43–52. [Google Scholar] [CrossRef]
- Chang, J.; Weigele, P.; King, J.; Chiu, W.; Jiang, W. Cryo-EM Asymmetric Reconstruction of Bacteriophage P22 Reveals Organization of its DNA Packaging and Infecting Machinery. Structure 2006, 14, 1073–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varki, A.; Cummings, R.D.; Aebi, M.; Packer, N.H.; Seeberger, P.H.; Esko, J.D.; Stanley, P.; Hart, G.; Darvill, A.; Kinoshita, T.; et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 25, 1323–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knirel, Y.A.; Sun, Q.; Senchenkova, S.N.; Perepelov, A.V.; Shashkov, A.S.; Xu, J. O-Antigen modifications providing antigenic diversity of Shigella flexneri and underlying genetic mechanisms. Biochem. Mosc. 2015, 80, 901–914. [Google Scholar] [CrossRef] [PubMed]
- RKI—Salmonellen und Andere Bakterielle Enteritis-Erreger: Leistungen. Available online: https://www.rki.de/DE/Content/Infekt/NRZ/Salmonellen/leistungen/leistungen_node.html (accessed on 17 May 2018).
- Bélot, F.; Guerreiro, C.; Baleux, F.; Mulard, L.A. Synthesis of Two Linear PADRE Conjugates Bearing a Deca- or Pentadecasaccharide B Epitope as Potential Synthetic Vaccines against Shigella flexneri Serotype 2a Infection. Chem. Eur. J. 2005, 11, 1625–1635. [Google Scholar] [CrossRef] [PubMed]
- Andres, D.; Gohlke, U.; Broeker, N.K.; Schulze, S.; Rabsch, W.; Heinemann, U.; Barbirz, S.; Seckler, R. An essential serotype recognition pocket on phage P22 tailspike protein forces Salmonella enterica serovar Paratyphi A O-antigen fragments to bind as nonsolution conformers. Glycobiology 2013, 23, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Zaccheus, M.V.; Broeker, N.K.; Lundborg, M.; Uetrecht, C.; Barbirz, S.; Widmalm, G. Structural studies of the O-antigen polysaccharide from Escherichia coli TD2158 having O18 serogroup specificity and aspects of its interaction with the tailspike endoglycosidase of the infecting bacteriophage HK620. Carbohydr. Res. 2012, 357, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, V.E.V.; Di Nardo, G.; Catucci, G.; Sadeghi, S.J.; Gilardi, G. Fluorescence detection of ligand binding to labeled cytochrome P450BM3. Dalton Trans. 2012, 41, 2018–2025. [Google Scholar] [CrossRef] [PubMed]
- Baxa, U.; Steinbacher, S.; Miller, S.; Weintraub, A.; Huber, R.; Seckler, R. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys. J. 1996, 71, 2040–2048. [Google Scholar] [CrossRef]
- Li, Y.; Cao, B.; Liu, B.; Liu, D.; Gao, Q.; Peng, X.; Wu, J.; Bastin, D.A.; Feng, L.; Wang, L. Molecular detection of all 34 distinct O-antigen forms of Shigella. J. Med. Microbiol. 2009, 58, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Kondakova, A.N.; Vinogradov, E.V.; Shekht, M.E.; Markina, A.A.; Lindner, B.; L’vov, V.L.; Aparin, P.G.; Knirel, Y.A. Structure of the oligosaccharide region (core) of the lipopolysaccharides of Shigella flexneri types 2a and 5b. Russ. J. Bioorg. Chem. 2010, 36, 396–399. [Google Scholar] [CrossRef]
- Mann, E.; Ovchinnikova, O.G.; King, J.D.; Whitfield, C. Bacteriophage-mediated Glucosylation Can Modify Lipopolysaccharide O-Antigens Synthesized by an ATP-binding Cassette (ABC) Transporter-dependent Assembly Mechanism. J. Biol. Chem. 2015, 290, 25561–25570. [Google Scholar] [CrossRef] [PubMed]
- Gettins, P.G.W.; Fan, B.; Crews, B.C.; Turko, I.V.; Olson, S.T.; Streusand, V.J. Transmission of conformational change from the heparin binding site to the reactive center of antithrombin. Biochemistry 1993, 32, 8385–8389. [Google Scholar] [CrossRef] [PubMed]
- Simmons, D.A.R.; Romanowska, E. Structure and biology of Shigella flexneri O antigens. J. Med. Microbiol. 1987, 23, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Van der Woude, M.W. Phase variation: How to create and coordinate population diversity. Curr. Opin. Microbiol. 2011, 14, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Parent, K.N.; Erb, M.L.; Cardone, G.; Nguyen, K.; Gilcrease, E.B.; Porcek, N.B.; Pogliano, J.; Baker, T.S.; Casjens, S.R. OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Mol. Microbiol. 2014, 92, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Porcek, N.B.; Parent, K.N. Key Residues of S. flexneri OmpA Mediate Infection by Bacteriophage Sf6. J. Mol. Biol. 2015, 427, 1964–1976. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Barbirz, S.; Lipowsky, R.; Santer, M. Conformational Diversity of O-Antigen Polysaccharides of the Gram-Negative Bacterium Shigella flexneri Serotype Y. J. Phys. Chem. B 2014, 118, 2523–2534. [Google Scholar] [CrossRef] [PubMed]
- Broeker, N.; Kiele, F.; Casjens, S.; Gilcrease, E.; Thalhammer, A.; Koetz, J.; Barbirz, S. In Vitro Studies of Lipopolysaccharide-Mediated DNA Release of Podovirus HK620. Viruses 2018, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Leon-Velarde, C.G.; Happonen, L.; Pajunen, M.; Leskinen, K.; Kropinski, A.M.; Mattinen, L.; Rajtor, M.; Zur, J.; Smith, D.; Chen, S.; et al. Yersinia enterocolitica-Specific Infection by Bacteriophages TG1 and ϕR1-RT Is Dependent on Temperature-Regulated Expression of the Phage Host Receptor OmpF. Appl. Environ. Microbiol. 2016, 82, 5340–5353. [Google Scholar] [CrossRef] [PubMed]
- Carlin, N.I.A.; Wehler, T.; Lindberg, A.A. Shigella flexneri O-Antigen Epitopes: Chemical and Immunochemical Analyses Reveal That Epitopes of Type III and Group 6 Antigens Are Identical. Infect. Immun. 1986, 53, 110–115. [Google Scholar] [PubMed]
- Vulliez-Le Normand, B.; Saul, F.A.; Phalipon, A.; Belot, F.; Guerreiro, C.; Mulard, L.A.; Bentley, G.A. Structures of synthetic O-antigen fragments from serotype 2a Shigella flexneri in complex with a protective monoclonal antibody. Proc. Natl. Acad. Sci. USA 2008, 105, 9976–9981. [Google Scholar] [CrossRef] [PubMed]
- Vyas, N.K.; Vyas, M.N.; Chervenak, M.C.; Johnson, M.A.; Pinto, B.M.; Bundle, D.R.; Quiocho, F.A. Molecular Recognition of Oligosaccharide Epitopes by a Monoclonal Fab Specific for Shigella flexneri Y Lipopolysaccharide: X-ray Structures and Thermodynamics. Biochemistry 2002, 41, 13575–13586. [Google Scholar] [CrossRef] [PubMed]
- Theillet, F.-X.; Chassagne, P.; Delepierre, M.; Phalipon, A.; Mulard, L.A. Multidisciplinary Approaches to Study O-Antigen: Antibody Recognition in Support of the Development of Synthetic Carbohydrate-Based Enteric Vaccines. In Anticarbohydrate Antibodies; Springer: Vienna, Austria, 2012; pp. 1–36. ISBN 978-3-7091-0869-7. [Google Scholar]
- Phalipon, A.; Tanguy, M.; Grandjean, C.; Guerreiro, C.; Bélot, F.; Cohen, D.; Sansonetti, P.J.; Mulard, L.A. A Synthetic Carbohydrate-Protein Conjugate Vaccine Candidate against Shigella flexneri 2a Infection. J. Immunol. 2009, 182, 2241–2247. [Google Scholar] [CrossRef] [PubMed]
- Van der Put, R.M.F.; Kim, T.H.; Guerreiro, C.; Thouron, F.; Hoogerhout, P.; Sansonetti, P.J.; Westdijk, J.; Stork, M.; Phalipon, A.; Mulard, L.A. A Synthetic Carbohydrate Conjugate Vaccine Candidate against Shigellosis: Improved Bioconjugation and Impact of Alum on Immunogenicity. Bioconjug. Chem. 2016, 27, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Micoli, F.; Romano, M.R.; Tontini, M.; Cappelletti, E.; Gavini, M.; Proietti, D.; Rondini, S.; Swennen, E.; Santini, L.; Filippini, S.; et al. Development of a glycoconjugate vaccine to prevent meningitis in Africa caused by meningococcal serogroup X. Proc. Natl. Acad. Sci. USA 2013, 110, 19077–19082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kämpf, M.M.; Braun, M.; Sirena, D.; Ihssen, J.; Thöny-Meyer, L.; Ren, Q. In vivo production of a novel glycoconjugate vaccine against Shigella flexneri 2a in recombinant Escherichia coli: Identification of stimulating factors for in vivo glycosylation. Microb. Cell Factories 2015, 14. [Google Scholar] [CrossRef] [PubMed]
- Niebuhr, K.; Jouihri, N.; Allaoui, A.; Gounon, P.; Sansonetti, P.J.; Parsot, C. IpgD, a protein secreted by the type III secretion machinery of Shigella flexneri, is chaperoned by IpgE and implicated in entry focus formation. Mol. Microbiol. 2000, 38, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Parsot, C.; Sansonetti, P.J. Invasion and the pathogenesis of Shigella infections. Curr. Top. Microbiol. Immunol. 1996, 209, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Van den Beld, M.J.C.; Friedrich, A.W.; van Zanten, E.; Reubsaet, F.A.G.; Kooistra-Smid, M.A.M.D.; Rossen, J.W.A. Multicenter evaluation of molecular and culture-dependent diagnostics for Shigella species and Entero-invasive Escherichia coli in the Netherlands. J. Microbiol. Methods 2016, 131, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Loferer-Krößbacher, M.; Klima, J.; Psenner, R. Determination of Bacterial Cell Dry Mass by Transmission Electron Microscopy and Densitometric Image Analysis. Appl. Environ. Microbiol. 1998, 64, 688–694. [Google Scholar] [PubMed]
- Darveau, R.P.; Hancock, R.E. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 1983, 155, 831–838. [Google Scholar] [PubMed]
- Schoonbroodt, S.; Steukers, M.; Viswanathan, M.; Frans, N.; Timmermans, M.; Wehnert, A.; Nguyen, M.; Ladner, R.C.; Hoet, R.M. Engineering Antibody Heavy Chain CDR3 to Create a Phage Display Fab Library Rich in Antibodies That Bind Charged Carbohydrates. J. Immunol. 2008, 181, 6213–6221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malou, N.; Tran, T.-N.-N.; Nappez, C.; Signoli, M.; Forestier, C.L.; Castex, D.; Drancourt, M.; Raoult, D. Immuno-PCR—A New Tool for Paleomicrobiology: The Plague Paradigm. PLoS ONE 2012, 7, e31744. [Google Scholar] [CrossRef] [PubMed]


| Bacterial Isolate 1 | Anti-S. flexneri Monoclonal Antibody | PCR (wzx) 2 | |
|---|---|---|---|
| Group 3,4 (Y) | Type 2 | ||
| S. flexneri Y 99-2001 | + | − | + |
| S. flexneri Y 03-650 | + | − | + |
| S. flexneri 2a 03-6557 | + | + | + |
| S. flexneri 2a 08-7230 | + | + | - |
| Sf6 TSP E366A/D399A Cysteine Mutant | Labeling Efficiency % | NBD Fluorescence Amplitude Increase at 540 nm 1 % |
|---|---|---|
| V204C | 19.4 | −10 |
| S246C | 42.8 | 53 |
| T315C | 43.2 | −15 |
| N340C | 66.7 | 78 |
| Y400C | 48.3 | 25 |
| T443C | 63.3 | 12 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kunstmann, S.; Scheidt, T.; Buchwald, S.; Helm, A.; Mulard, L.A.; Fruth, A.; Barbirz, S. Bacteriophage Sf6 Tailspike Protein for Detection of Shigella flexneri Pathogens. Viruses 2018, 10, 431. https://doi.org/10.3390/v10080431
Kunstmann S, Scheidt T, Buchwald S, Helm A, Mulard LA, Fruth A, Barbirz S. Bacteriophage Sf6 Tailspike Protein for Detection of Shigella flexneri Pathogens. Viruses. 2018; 10(8):431. https://doi.org/10.3390/v10080431
Chicago/Turabian StyleKunstmann, Sonja, Tom Scheidt, Saskia Buchwald, Alexandra Helm, Laurence A. Mulard, Angelika Fruth, and Stefanie Barbirz. 2018. "Bacteriophage Sf6 Tailspike Protein for Detection of Shigella flexneri Pathogens" Viruses 10, no. 8: 431. https://doi.org/10.3390/v10080431





