Relative Contribution of Cellular Complement Inhibitors CD59, CD46, and CD55 to Parainfluenza Virus 5 Inhibition of Complement-Mediated Neutralization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Ultracentrifugation and Western Blotting
2.3. Neutralization Assay
2.4. Flow Cytometric Analysis
2.5. Reverse Transcription and Real Time PCR
2.6. C9 Polymerization Assay
2.7. Complement Deposition on Infected Cells
3. Results
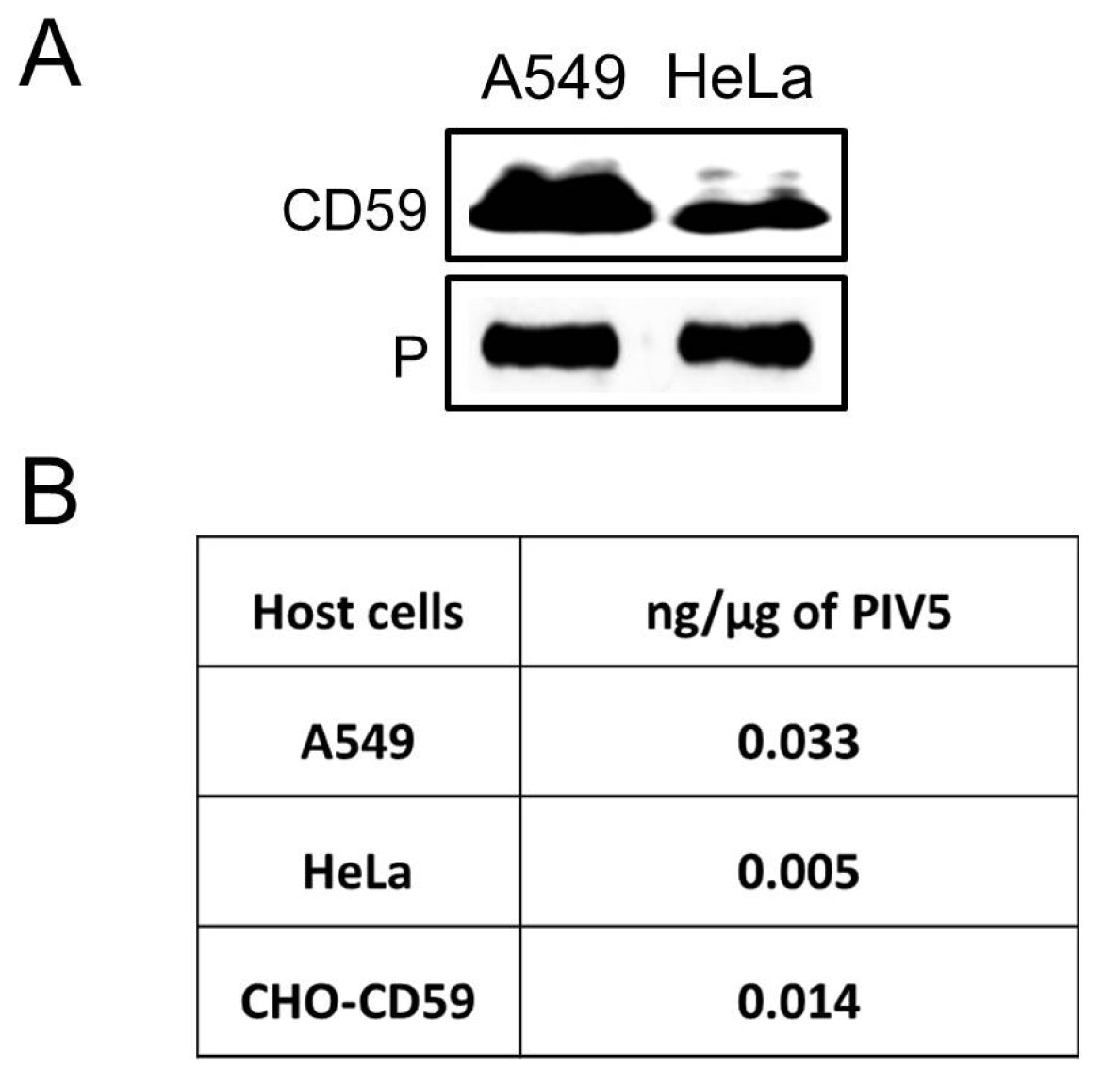
3.1. CD59 Is Associated with PIV5 Virions
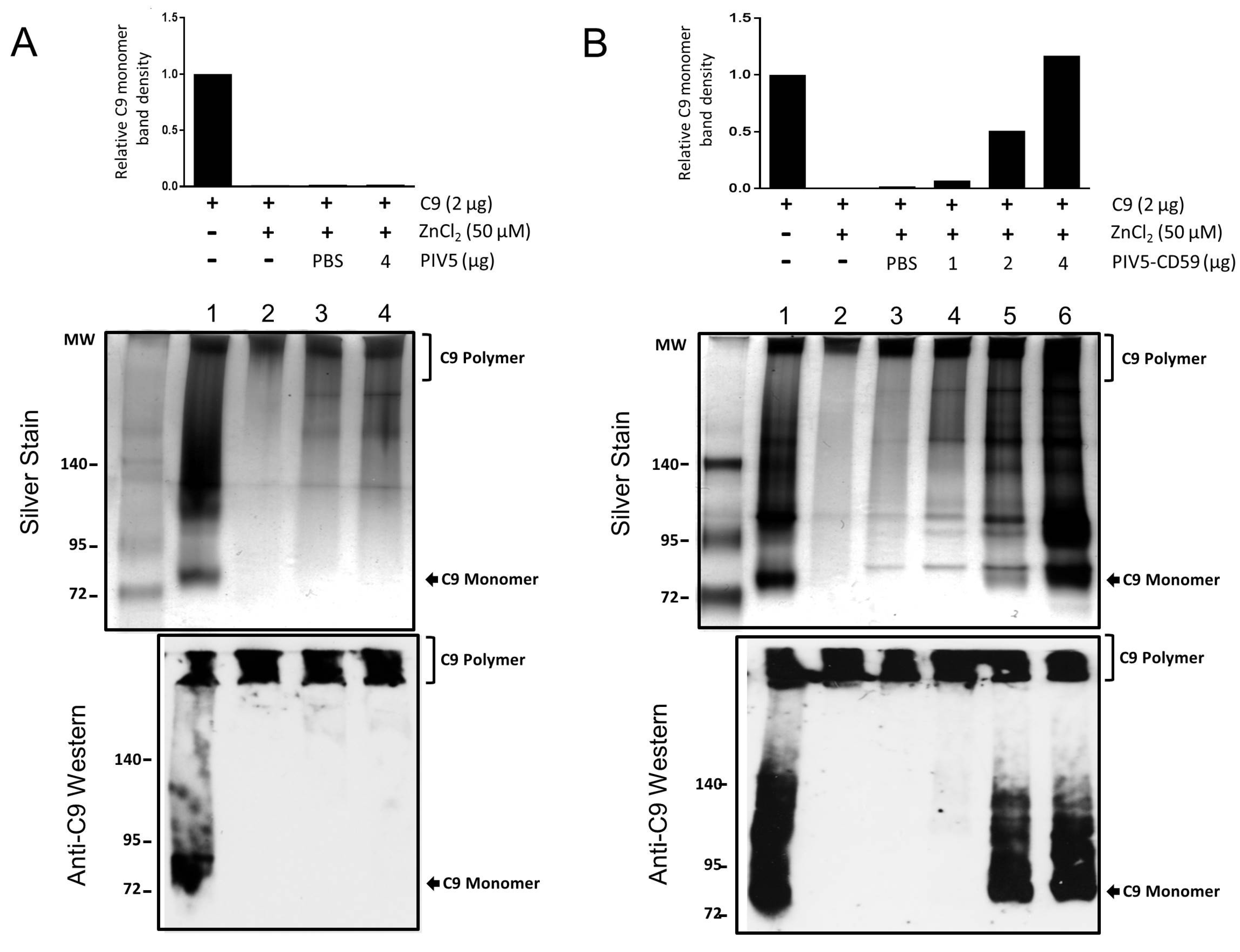
3.2. CD59 Contained in PIV5-CD59 Inhibits C9 Polymerization
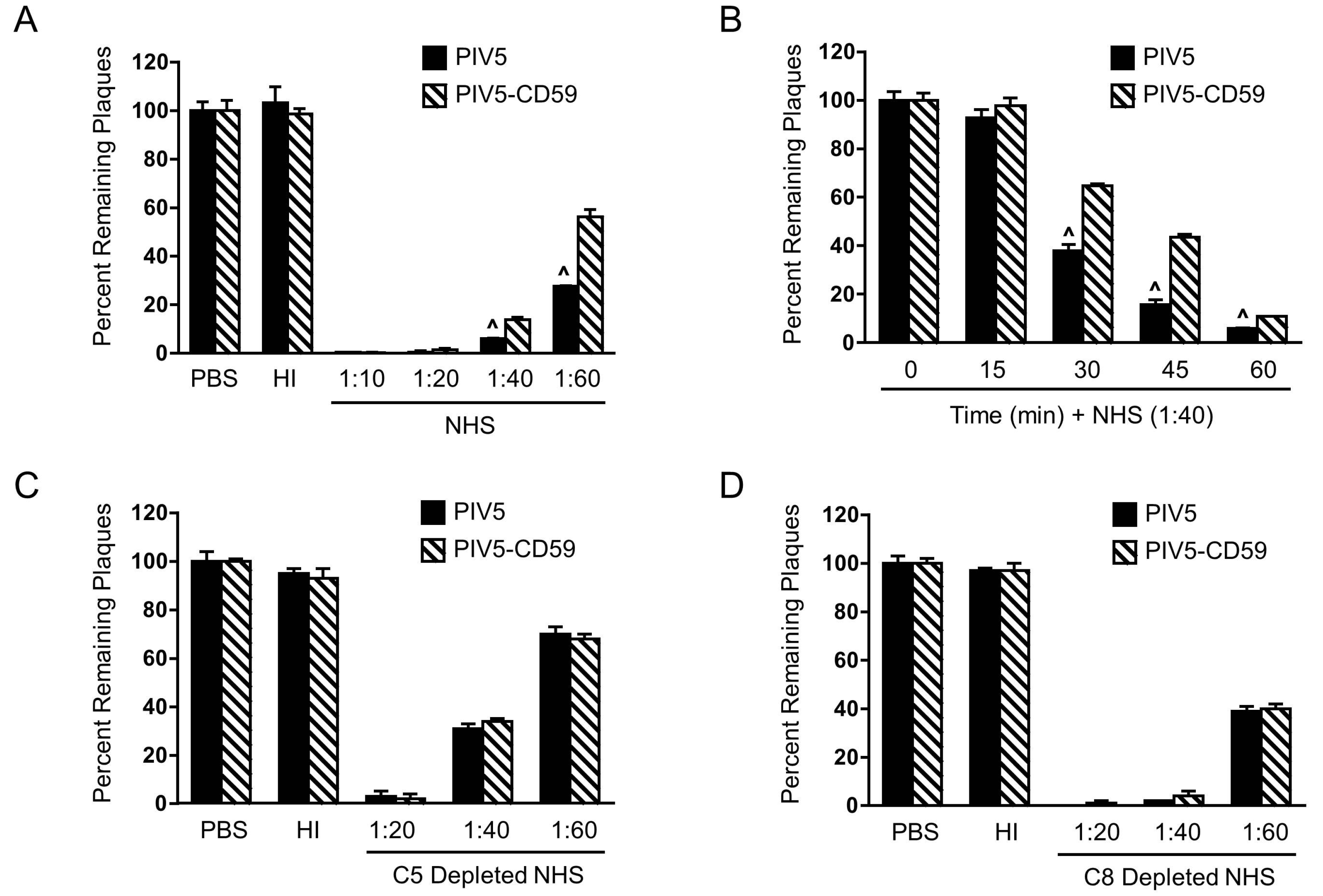
3.3. PIV5 Derived from CD59-Expressing Cells Has Increased Resistance to Complement-Mediated Neutralization In Vitro
3.4. Relative Contribution of CD59, CD46, and CD55 to PIV5 Complement-Mediated Neutralization
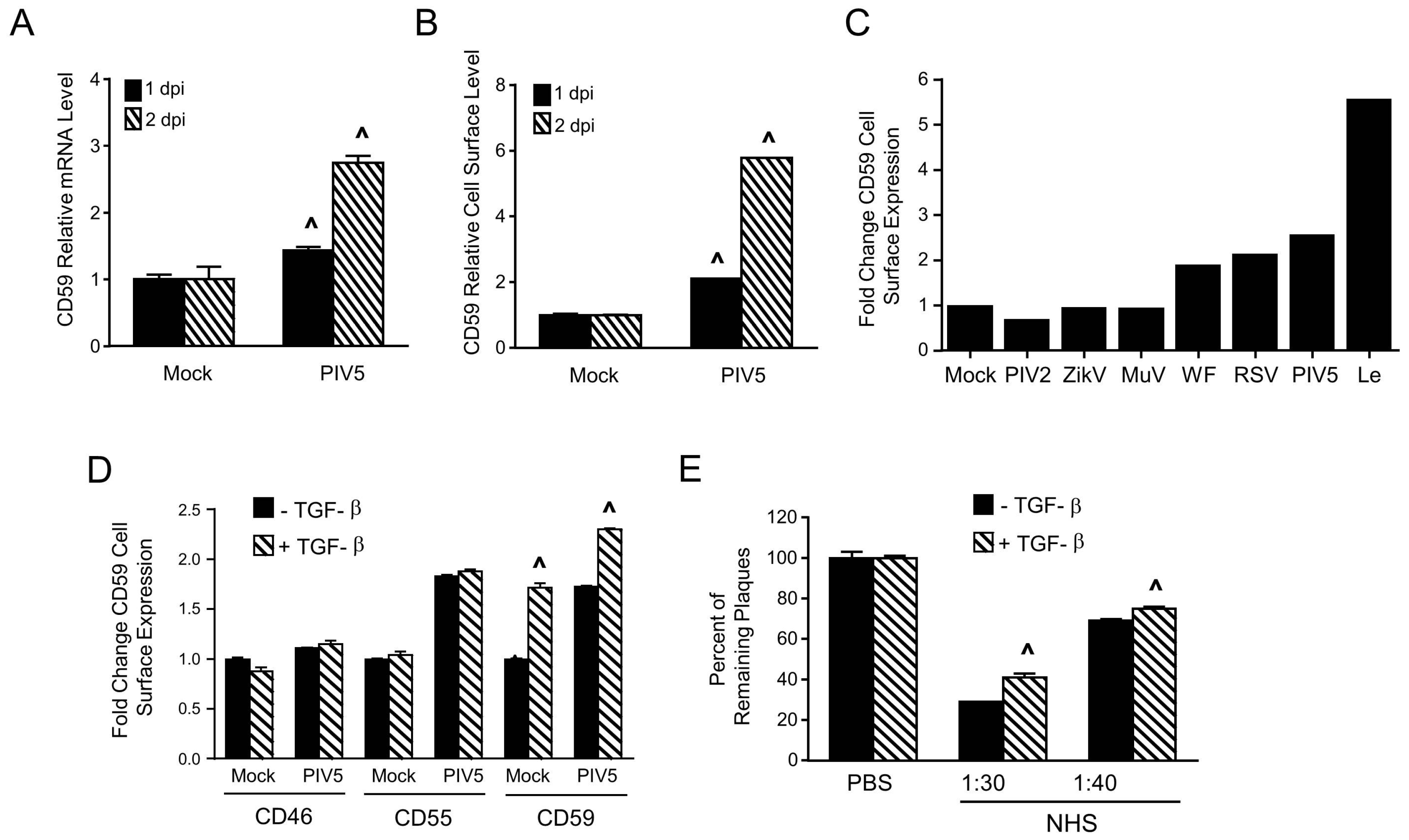
3.5. PIV5 Infection Upregulates Cell Surface CD59 and Confers Virions with Increased Resistance to Complement-Mediated Neutralization
4. Discussion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Blue, C.E.; Spiller, O.B.; Blackbourn, D.J. The relevance of complement to virus biology. Virology 2004, 319, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Carroll, M.C. The complement system in regulation of adaptive immunity. Nat. Immunol. 2004, 5, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Kemper, C.; Atkinson, J.P. T-cell regulation: With complements from innate immunity. Nat. Rev. Immunol. 2007, 7, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Mehlhop, E.; Diamond, M.S. Protective immune responses against West Nile virus are primed by distinct complement activation pathways. J. Exp. Med. 2006, 203, 1371–1381. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.E.; Fraser, R.J.; Smith, P.N.; Mahalingam, S.; Heise, M.T. Complement contributes to inflammatory tissue destruction in a mouse model of Ross River virus-induced disease. J. Virol. 2007, 81, 5132–5143. [Google Scholar] [CrossRef] [PubMed]
- Delgado, M.F.; Polack, F.P. Involvement of antibody, complement and cellular immunity in the pathogenesis of enhanced respiratory syncytial virus disease. Expert Rev. Vaccines 2004, 3, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Stoermer, K.A.; Morrison, T.E. Complement and viral pathogenesis. Virology 2011, 411, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.B.; Aguilar, H.C.; Lee, B.; Parks, G.D. Interactions of human complement with virus particles containing the Nipah virus glycoproteins. J. Virol. 2011, 85, 5940–5948. [Google Scholar] [CrossRef] [PubMed]
- Bergmann-Leitner, E.S.; Leitner, W.W.; Tsokos, G.C. Complement 3d: From molecular adjuvant to target of immune escape mechanisms. Clin. Immunol. 2006, 121, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Schauber-Plewa, C.; Simmons, A.; Tuerk, M.J.; Pacheco, C.D.; Veres, G. Complement regulatory proteins are incorporated into lentiviral vectors and protect particles against complement inactivation. Gene Ther. 2005, 12, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Markiewski, M.M.; Lambris, J.D. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am. J. Pathol. 2007, 171, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Roozendaal, R.; Carroll, M.C. Emerging patterns in complement-mediated pathogen recognition. Cell 2006, 125, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Kerr, M.A. The human complement system: Assembly of the classical pathway C3 convertase. Biochem. J. 1980, 189, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.P.; Liszewski, M.K.; Richards, A.; Kavanagh, D.; Moulton, E.A. Hemolytic uremic syndrome: An example of insufficient complement regulation on self-tissue. Ann. N. Y. Acad. Sci. 2005, 1056, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Pangburn, M.K.; Schreiber, R.D.; Muller-Eberhard, H.J. Deficiency of an erythrocyte membrane protein with complement regulatory activity in paroxysmal nocturnal hemoglobinuria. Proc. Natl. Acad. Sci. USA 1983, 80, 5430–5434. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.D.; Song, W.C. Membrane complement regulatory proteins. Clin. Immunol. 2006, 118, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Meri, S.; Wldmann, H.; Lachmann, P.J. Distribution of protectin (CD59), a complement membrane attack inhibitor, in normal human tissues. Lab. Investig. 1991, 65, 532–537. [Google Scholar] [PubMed]
- Shan, C.; Zhang, S.; Cui, W.; You, X.; Kong, G.; Du, Y.; Qiu, L.; Ye, L.; Zhang, X. Hepatitis B virus X protein activates CD59 involving DNA binding and let-7i in protection of hepatoma and hepatic cells from complement attack. Carcinogenesis 2011, 32, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Morgan, B.P. Regulation of the complement membrane attack pathway. Crit. Rev. Immunol. 1999, 19, 173–198. [Google Scholar] [CrossRef] [PubMed]
- Meri, S.; Morgan, B.P.; Davies, A.; Daniels, R.H.; Olavesen, M.G.; Waldmann, H.; Lachmann, P.J. Human protectin (CD59), an 18,000–20,000 MW complement lysis restricting factor, inhibits C5b-8 catalysed insertion of C9 into lipid bilayers. Immunology 1990, 71, 1–9. [Google Scholar] [PubMed]
- Rollins, S.A.; Sims, P.J. The complement-inhibitory activity of CD59 resides in its capacity to block incorporation of C9 into membrane C5b-9. J. Immunol. 1990, 144, 3478–3483. [Google Scholar] [PubMed]
- Isaacs, S.N.; Kotwal, G.J.; Moss, B. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc. Natl. Acad. Sci. USA 1992, 89, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.M.; Liszewski, M.K.; Nybakken, G.; Davis, A.E.; Townsend, R.R.; Fremont, D.H.; Atkinson, J.P.; Diamond, M.S. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc. Natl. Acad. Sci. USA 2006, 103, 19111–19116. [Google Scholar] [CrossRef] [PubMed]
- Cummings, K.L.; Waggoner, S.N.; Tacke, R.; Hahn, Y.S. Role of complement in immune regulation and its exploitation by virus. Viral Immunol. 2007, 20, 505–524. [Google Scholar] [CrossRef] [PubMed]
- Saifuddin, M.; Hedayati, T.; Atkinson, J.P.; Holguin, M.H.; Parker, C.J.; Spear, G.T. Human immunodeficiency virus type 1 incorporates both glycosyl phosphatidyinositor-anchored CD55 and CD59 and integral membrane CD46 at levels that protect from complement mediated destruction. J. Gen. Virol. 1997, 78, 1907–1911. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.L.; Stone, K.L.; Colangelo, C.M.; Gulcicek, E.E.; Palese, P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008, 4, e1000085. [Google Scholar] [CrossRef] [PubMed]
- Stoiber, H.; Pinter, C.; Siccardi, A.G.; Clivio, A.; Dierich, M.P. Efficient destruction of human immunodeficiency virus in human serum by inhibiting the protective action of complement factor H and decay accelerating factor (DAF, CD55). J. Exp. Med. 1996, 183, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Vanderplasschen, A.; Mathew, E.; Hollinshead, M.; Sim, R.B.; Smith, G.L. Extracellular enveloped vaccinia virus is resistant to complement because of incorporation of host complement control proteins into its envelope. Proc. Natl. Acad. Sci. USA 1998, 95, 7544–7549. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, R.L.; Wolinsky, J.S.; Winkelstein, J.A. Activation of the alternative complement pathway by mumps infected cells: Relationship to viral neuraminidase activity. Arch. Virol. 1986, 87, 181–190. [Google Scholar] [CrossRef] [PubMed]
- McSharry, J.J.; Pickering, R.J.; Caliguiri, L.A. Activation of the alternative complement pathway by enveloped viruses containing limited amounts of sialic acid. Virology 1981, 114, 507–515. [Google Scholar] [CrossRef]
- Johnson, J.B.; Grant, K.; Parks, G.D. The paramyxoviruses simian virus 5 and mumps virus recruit host cell CD46 to evade complement-mediated neutralization. J. Virol. 2009, 83, 7602–7611. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.B.; Lyles, D.S.; Alexander-Miller, M.A.; Parks, G.D. Virion-associated CD55 is more potent than CD46 in mediating resistance of mumps virus and VSV to neutralization. J. Virol. 2012, 86, 9929–9940. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Johnson, J.B.; Parks, G.D. Parainfluenza virus 5 upregulates CD55 expression to produce virions with enhanced resistance to complement-mediated neutralization. Virology 2016, 497, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Young, V.A.; Dillon, P.J.; Parks, G.D. Variants of the paramyxovirus Simian virus 5 with accelerated or delayed viral gene expression activate proinflammatory cytokine synthesis. Virology 2006, 350, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Manuse, M.J.; Parks, G.D. Role for the paramyxovirus genomic promoter in limiting host cell antiviral responses and cell killing. J. Virol. 2009, 83, 9057–9067. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.B.; Capraro, G.A.; Parks, G.D. Differential mechanisms of complement mediated neutralization of the closely related paramyxoviruses simian virus 5 and mumps virus. Virology 2008, 376, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kakinami, C.; Li, Q.; Yang, B.; Li, H. Human apolipoprotein A-I is associated with dengue virus and enhances virus infection through SR-BI. PLoS ONE 2013, 8, e70390. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.N.; da Silva, E.M.; Allonso, D.; Coelho, D.R.; Andrade, I.D.; de Medeiros, L.N.; Menezes, J.L.; Barbosa, A.S.; Mohana-Borges, R. Inhibition of the Membrane Attack Complex by Dengue Virus NS1 through Interaction with Vitronectin and Terminal Complement Proteins. J. Virol. 2016, 90, 9570–9581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, J.; Wei, J.; Yang, Y.; Chen, X.; Zhao, X.; Gu, Y.; Cui, S.; Zhu, X. Trichinella spiralis paramyosin binds to C8 and C9 and protects the tissue-dwelling nematode from being attacked by host complement. PLoS Negl. Trop. Dis. 2011, 5, e1225. [Google Scholar] [CrossRef] [PubMed]
- Brasoveanu, L.I.; Altomonte, M.; Fonsatti, E.; Colizzi, F.; Coral, S.; Nicotra, M.R.; Cattarossi, I.; Cattelan, A.; Natali, P.G.; Maio, M. Levels of cell membrane CD59 regulate the extent of complement-mediated lysis of human melanoma cells. Lab. Investig. 1996, 74, 33–42. [Google Scholar] [PubMed]
- Jarvis, G.A.; Li, J.; Hakulinen, J.; Brady, K.A.; Nordling, S.; Dahita, R.; Meri, S. Expression and function of the complement membrane attack complex inhibitor protectin (CD59) in human prostate cancer. Int. J. Cancer 1997, 71, 1049–1055. [Google Scholar] [CrossRef]
- Chen, S.; Caragine, T.; Cheung, N.K.; Tomlinson, S. CD59 expressed on a tumor cell surface modulates decay-accelerating factor expression and enhances tumor growth in a rat model of human neuroblastoma. Cancer Res. 2000, 60, 3013–3018. [Google Scholar] [PubMed]
- Coral, S.; Fonsatti, E.; Sigalotti, L.; De Nardo, C.; Visintin, A.; Nardi, G.; Colizzi, F.; Colombo, M.P.; Romano, G.; Altomonte, M.; et al. Overexpression of protectin (CD59) down-modulates the susceptibility of human melanoma cells to homologous complement. J. Cell. Physiol. 2000, 185, 317–323. [Google Scholar] [CrossRef]
- Jurianz, K.; Ziegler, S.; Garcia-Schüler, H.; Kraus, S.; Bohana-Kashtan, O.; Fishelson, Z.; Kirschfink, M. Complement resistance of tumor cells: Basal and induced mechanisms. Mol. Immunol. 1999, 36, 929–939. [Google Scholar] [CrossRef]
- Goswami, M.T.; Reka, A.K.; Kurapati, H.; Kaza, V.; Chen, J.; Standiford, T.J.; Keshamouni, V.G. Regulation of complement-dependent cytotoxicity by TGF-β-induced epithelial–mesenchymal transition. Oncogene 2016, 35, 1888–1898. [Google Scholar] [CrossRef] [PubMed]
- Song, W.C. Membrane complement regulatory proteins in autoimmune and inflammatory tissue injury. Curr. Dir. Autoimmun. 2004, 7, 181–199. [Google Scholar] [PubMed]
- Takemoto, M.; Yamanishi, K.; Mori, Y. Human herpesvirus 7 infection increases the expression levels of CD46 and CD59 in target cells. J. Gen. Virol. 2007, 88, 1415–1422. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Parks, G.D. Relative Contribution of Cellular Complement Inhibitors CD59, CD46, and CD55 to Parainfluenza Virus 5 Inhibition of Complement-Mediated Neutralization. Viruses 2018, 10, 219. https://doi.org/10.3390/v10050219
Li Y, Parks GD. Relative Contribution of Cellular Complement Inhibitors CD59, CD46, and CD55 to Parainfluenza Virus 5 Inhibition of Complement-Mediated Neutralization. Viruses. 2018; 10(5):219. https://doi.org/10.3390/v10050219
Chicago/Turabian StyleLi, Yujia, and Griffith D. Parks. 2018. "Relative Contribution of Cellular Complement Inhibitors CD59, CD46, and CD55 to Parainfluenza Virus 5 Inhibition of Complement-Mediated Neutralization" Viruses 10, no. 5: 219. https://doi.org/10.3390/v10050219



