High-Throughput Sequencing Shows High Fungal Diversity and Community Segregation in the Rhizospheres of Container-Grown Conifer Seedlings
Abstract
:1. Introduction
2. Experimental Section
2.1. Seedling Materials and Sampling
2.2. DNA Work and Bioinformatics
2.3. Statistical Analyses
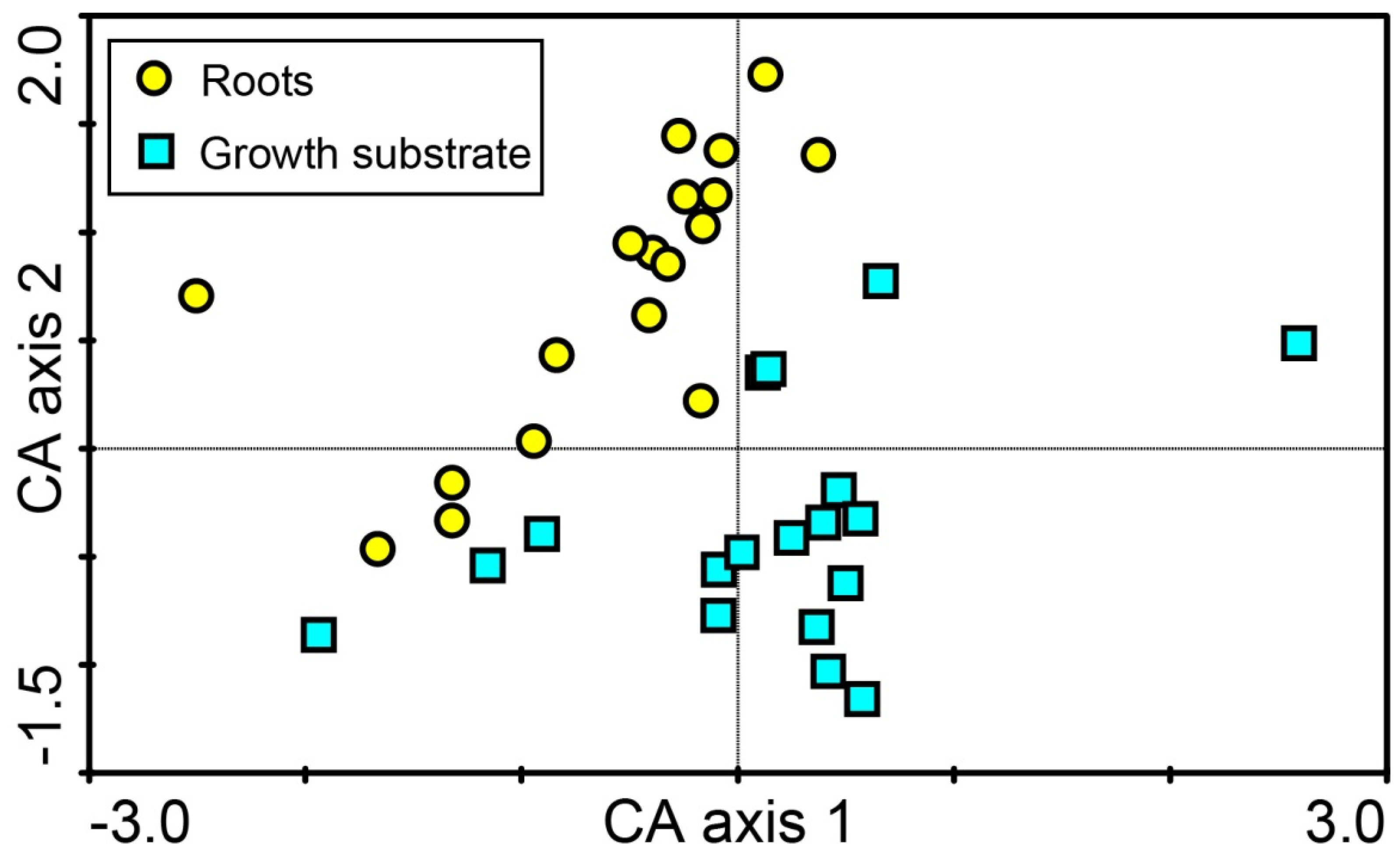
3. Results
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgements
Author Contributions
Conflicts of Interest
References
- Stenlid, J.; Oliva, J.; Boberg, J.B.; Hopkins, A.J.M. Emerging diseases in european forest ecosystems and responses in society. Forests 2011, 2, 486–504. [Google Scholar] [CrossRef]
- Wingfield, M.J.; Brockerhoff, E.G.; Wingfield, B.D.; Slippers, B. Planted forest health: The need for a global strategy. Science 2015, 349, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Queloz, V.; Grünig, C.R.; Berndt, R.; Kowalski, T.; Sieber, T.N.; Holdenrieder, O. Cryptic speciation in Hymenoscyphus albidus. For. Pathol. 2011, 41, 133–142. [Google Scholar] [CrossRef]
- Husson, C.; Scala, B.; Cael, O.; Frey, P.; Feau, N.; Ioos, R.; Marcais, B. Chalara fraxinea is an invasive pathogen in France. Eur. J. Plant Pathol. 2011, 130, 311–324. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Hosoya, T.; Baral, H.O.; Hosaka, K.; Kakishima, M. Hymenoscyphus pseudoalbidus, the correct name for Lambertella albida reported from Japan. Mycotaxon 2012, 122, 25–41. [Google Scholar] [CrossRef]
- Zheng, H.-D.; Zhuang, W.-Y. Hymenoscyphus albidoides sp. nov. and H. pseudoalbidus from China. Mycol. Prog. 2014, 13, 625–638. [Google Scholar] [CrossRef]
- Pautasso, M.; Aas, G.; Queloz, V.; Holdenrieder, O. European ash (Fraxinus excelsior) dieback—A conservation biology challenge. Biol. Conserv. 2013, 158, 37–49. [Google Scholar] [CrossRef]
- Bakys, R.; Vasaitis, R.; Barklund, P.; Thomsen, I.; Stenlid, J. Occurrence and pathogenicity of fungi in necrotic and non-symptomatic shoots of declining common ash (Fraxinus excelsior) in Sweden. Eur. J. For. Res. 2009, 128, 51–60. [Google Scholar] [CrossRef]
- Grunwald, N.J.; Goss, E.M.; Press, C.M. Phytophthora ramorum: A pathogen with a remarkably wide host range causing sudden oak death on oaks and ramorum blight on woody ornamentals. Mol. Plant Pathol. 2008, 9, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Ivors, K.; Garbelotto, M.; Vries, I.D.E.; Ruyter-Spira, C.; Hekkert, B.T.; Rosenzweig, N.; Bonants, P. Microsatellite markers identify three lineages of Phytophthora ramorum in US nurseries, yet single lineages in US forest and European nursery populations. Mol. Ecol. 2006, 15, 1493–1505. [Google Scholar] [CrossRef] [PubMed]
- Koch, F.H.; Smith, W.D. Mapping Sudden Oak Death Risk Nationally Using Host, Climate, and Pathways Data. In Proceedings of the Sudden Oak Death Third Science Symposium Santa Rosa, CA, USA, 5–9 March 2007; pp. 279–287.
- Jung, T.; Blaschke, M. Phytophthora root and collar rot of alders in Bavaria: Distribution, modes of spread and possible management strategies. Plant Pathol. 2004, 53, 197–208. [Google Scholar] [CrossRef]
- Menkis, A.; Vasiliauskas, R.; Taylor, A.F.S.; Stenström, E.; Stenlid, J.; Finlay, R. Fungi in decayed roots of conifer seedlings from forest nurseries, afforested clearcuts and abandoned farmland. Plant Pathol. 2006, 55, 117–129. [Google Scholar] [CrossRef]
- Menkis, A.; Vasaitis, R. Fungi in roots of nursery grown Pinus sylvestris: Ectomycorrhizal colonisation, genetic diversity and spatial distribution. Microb. Ecol. 2011, 61, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Menkis, A.; Vasiliauskas, R.; Taylor, A.F.S.; Stenlid, J.; Finlay, R. Fungal communities in mycorrhizal roots of conifer seedlings in forest nurseries under different cultivation systems, assessed by morphotyping, direct sequencing and mycelial isolation. Mycorrhiza 2005, 16, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Menkis, A.; Vasiliauskas, R.; Taylor, A.F.S.; Stenlid, J.; Finlay, R. Afforestation of abandoned farmland with conifer seedlings inoculated with three ectomycorrhizal fungi—Impact on plant performance and ectomycorrhizal community. Mycorrhiza 2007, 17, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 2nd ed.; Academic Press: London, UK, 1997; p. 605. [Google Scholar]
- Saikkonen, K.; Faeth, S.H.; Helander, M.; Sullivan, T.J. Fungal endophytes: A continuum of interactions with host plants. Annu. Rev. Ecol. Syst. 1998, 29, 319–343. [Google Scholar] [CrossRef]
- Sieber, T.N. Fungal root endophytes. In Plant Roots: The Hidden Half, 3rd ed.; Wasel, Y., Eshel, A., Kafkafi, U., Eds.; Marcel Dekker: New York, NY, USA, 2002; pp. 887–917. [Google Scholar]
- Findlay, J.A.; Li, G.Q.; Miller, J.D.; Womiloju, T.O. Insect toxins from spruce endophytes. Can. J. Chem. 2004, 81, 284–292. [Google Scholar] [CrossRef]
- Broberg, A.; Menkis, A.; Vasiliauskas, R. Kutznerides 1–4, depsipeptides from the actinomycete Kutzneria sp. 744 inhabiting mycorrhizal roots of Picea abies seedlings. J. Nat. Prod. 2006, 69, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Stenström, E.; Ndobe, N.E.; Jonsson, M.; Stenlid, J.; Menkis, A. Root associated fungi of healthy-looking Pinus sylvestris and Picea abies seedlings in Swedish forest nurseries. Scand. J. For. Res. 2014, 29, 12–21. [Google Scholar] [CrossRef]
- Beyer-Ericson, L.; Damm, E.; Unestam, T. An overview of root dieback and its causes in Swedish forest nurseries. Eur. J. For. Pathol. 1991, 21, 439–443. [Google Scholar] [CrossRef]
- Unestam, T.; Beyer-Ericson, L.; Strand, M. Involvement of Cylindrocarpon destructans in root death of Pinus sylvestris seedlings: Pathogenic behaviour and predisposing factors. Scand. J. For. Res. 1989, 4, 521–535. [Google Scholar] [CrossRef]
- Menkis, A.; Burokienė, D.; Gaitnieks, T.; Uotila, A.; Johannesson, H.; Rosling, A.; Finlay, R.D.; Stenlid, J.; Vasaitis, R. Occurrence and impact of the root-rot biocontrol agent Phlebiopsis gigantea on soil fungal communities in Picea abies forests of northern Europe. FEMS Microbiol. Ecol. 2012, 81, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Wennström, U.; Johansson, K.; Lindström, A.; Stattin, E. Produktion av frö och plantor [Production of Seeds and Seedlings]; Skogsskötselserien nr 2; Skogsstyrelsen: Jönköping, Sweden, 2008. (In Swedish) [Google Scholar]
- Beyer-Ericson, L. Svampsjukdomar i Skogsplantskolor [Fungal Diseases in Forest Nurseries]; SLU: Uppsala, Sweden, 1990. (In Swedish) [Google Scholar]
- Ihrmark, K.; Bodeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandstrom-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press, Inc.: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Center for Microbial Ecology, Michigan State University. The Ribosomal Database Project. Available online: https://pyro.cme.msu.edu/index.jsp (accessed on 3 November 2015).
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Magurran, A.E. Ecological Diversity and Its Measurement; Princeton University Press: Princeton, NJ, USA, 1988; p. 192. [Google Scholar]
- Sokal, R.R.; Rohlf, F.J. Biometry: The Principles and Practice of Statistics in Biological Research, 3rd ed.; W.H. Freeman and Co: New York, NY, USA, 1995; p. 887. [Google Scholar]
- Minitab® Inc. Minitab Statistical Software; Release 15.1; Minitab Inc.: State College, PA, USA, 2003. [Google Scholar]
- Chalmers, N.; Parker, P. Fieldwork and Statistics for Ecological Projects, 2nd ed.; The Open University: Dorchester, UK, 1989; p. 108. [Google Scholar]
- Fowler, J.; Cohen, L.; Jarvis, P. Practical Statistics for Field Biology, 2nd ed.; Wiley: Chichester, UK, 1998; p. 259. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Ter Braak, C.J.F.; Smilauer, P. Canoco Reference Manual and User’s Guide to Canoco for Windows: Software for Canonical Community Ordination; Version 4; Microcomputer Power: Ithaca, NY, USA, 1998; p. 351. [Google Scholar]
- Kernaghan, G.; Sigler, L.; Khasa, D. Mycorrhizal and root endophytic fungi of containerized Picea glauca seedlings assessed by rDNA sequence analysis. Microb. Ecol. 2003, 45, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Lilja, A.; Rikala, R. Effect of uninucleate Rhizoctonia on the survival of outplanted Scots pine and Norway spruce seedlings. For. Pathol. 2000, 30, 109–115. [Google Scholar] [CrossRef]
- Voříšková, J.; Brabcová, V.; Cajthaml, T.; Baldrian, P. Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytol. 2014, 201, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Rudawska, M.; Leski, T.; Trocha, L.K.; Gornowicz, R. Ectomycorrhizal status of Norway spruce seedlings from bare-root forest nurseries. For. Ecol. Manag. 2006, 236, 375–384. [Google Scholar] [CrossRef]
- Kirk, P.M.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Dictionary of the Fungi; CABI: Wallingford, UK, 2008; p. 310. [Google Scholar]
- Klavina, D.; Gaitnieks, T.; Menkis, A. Survival, growth and ectomycorrhizal community development of container- and bare-root grown Pinus sylvestris and Picea abies seedlings outplanted on a forest clear-cut. Balt. For. 2013, 19, 39–49. [Google Scholar]
- Kurtzman, C.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study, 5th ed.; Elsevier: Burlington, VT, USA, 2011. [Google Scholar]
- Yurkov, A.M.; Schafer, A.M.; Begerow, D. Leucosporidium drummii sp. nov., a member of the Microbotryomycetes isolated from soil. Int. J. Syst. Evolut. Microbiol. 2012, 62, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.L.; Currah, R.S. Atradidymella muscivora gen. et sp. nov. (Pleosporales) and its anamorph Phoma muscivora sp. nov.: A new pleomorphic pathogen of boreal bryophytes. Am. J. Bot. 2009, 96, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Capieau, K. Biological Control of Grey Mould in Swedish Forest Nurseries. Ph.D. Thesis, Swedish University of Agricultural Sciences, Uppsala, Sweden, 2004. [Google Scholar]
- Näsholm, T.; Ekblad, A.; Nordin, A.; Giesler, R.; Högberg, M.; Högberg, P. Boreal forest plants take up organic nitrogen. Nature 1998, 392, 914–916. [Google Scholar] [CrossRef]
- Lindahl, B.; Stenlid, J.; Olsson, S.; Finlay, R. Translocation of P-32 between interacting mycelia of a wood-decomposing fungus and ectomycorrhizal fungi in microcosm systems. New Phytol. 1999, 144, 183–193. [Google Scholar] [CrossRef]
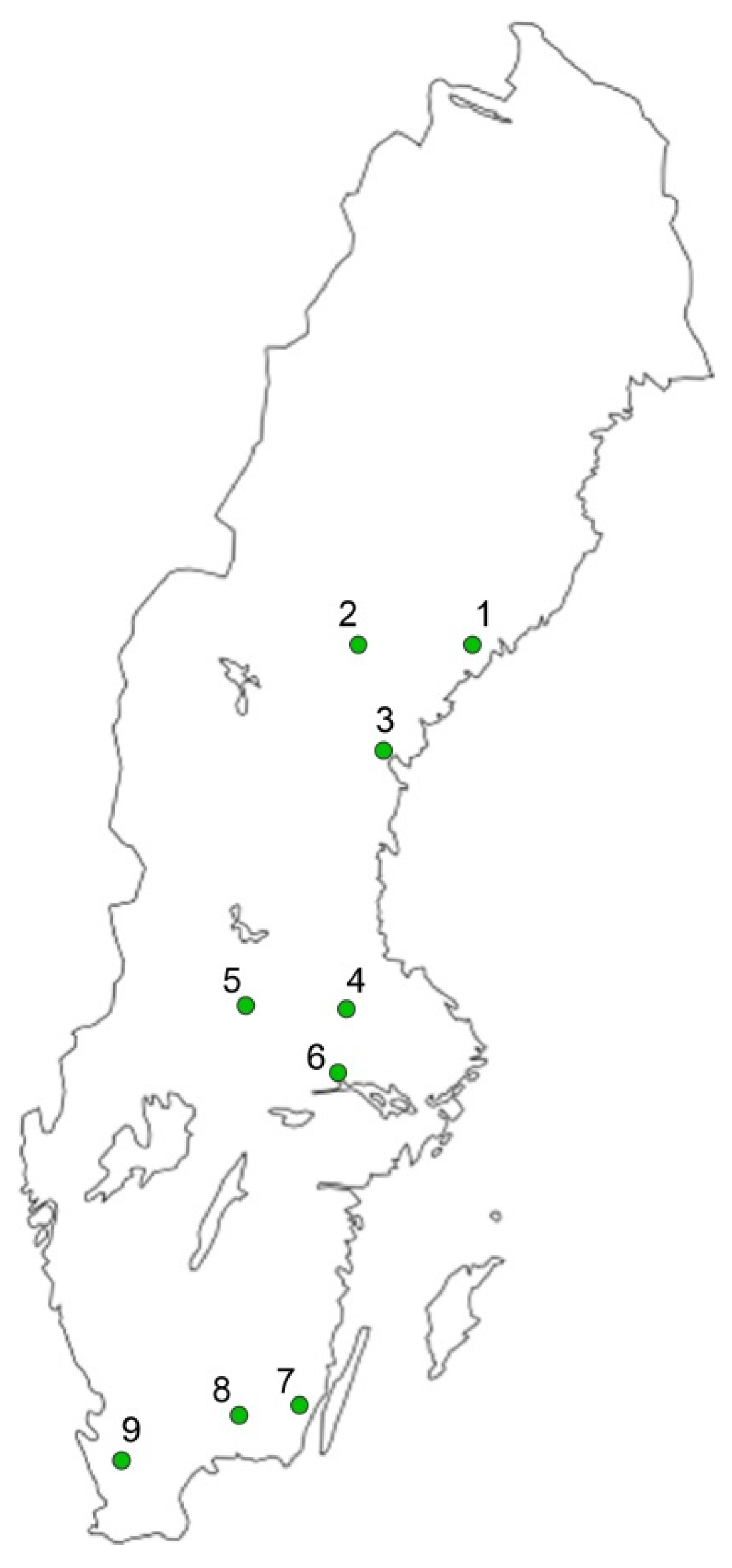
| No. | Nursery | Location | Pinus sylvestris a | Picea abies a | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Roots | Sequences | Taxa | Peat | Sequences | Taxa | Roots | Sequences | Taxa | Peat | Sequences | Taxa | |||
| 1 | Gideå | N63°29′, E18°58′ | 100 | 6528 | 59 | 100 | 7246 | 70 | 100 | 2291 | 46 | 100 | 5221 | 76 |
| 2 | Kilåmon | N63°28′, E16°40′ | 100 | 2242 | 72 | 100 | 6988 | 100 | 100 | 2599 | 35 | 100 | 3390 | 83 |
| 3 | Bogrundet | N62°31′, E17°18′ | 100 | 1548 | 52 | 100 | 2122 | 96 | 100 | 664 | 41 | 100 | 4028 | 126 |
| 4 | Nässja | N60°16′, E16°42′ | 100 | 637 | 30 | 100 | 3813 | 48 | 100 | 1201 | 36 | 100 | 5363 | 92 |
| 5 | Lugnet | N59°41′, E16°36′ | 100 | 1171 | 50 | 100 | 7261 | 127 | 100 | 1339 | 46 | 100 | 7941 | 124 |
| 6 | Stakheden | N60°16′, E14°57′ | 100 | 712 | 23 | 100 | 8005 | 120 | 100 | 1964 | 39 | 100 | 9836 | 110 |
| 7 | Trekanten | N56°41′, E16°06′ | 100 | 8113 | 100 | 100 | 10392 | 149 | 100 | 9209 | 75 | 100 | 8351 | 139 |
| 8 | Flåboda | N56°34′, E15°08′ | 100 | 1330 | 40 | 100 | 5611 | 92 | 100 | 686 | 53 | 100 | 3131 | 90 |
| 9 | Kolleberga | N56°03′, E13°15′ | 100 | 2763 | 92 | 100 | 9357 | 164 | 100 | 438 | 33 | 100 | 7668 | 197 |
| Total | 900 | 25044 | 237 | 900 | 60795 | 406 | 900 | 20391 | 159 | 900 | 54929 | 442 | ||
| Fungal taxa | Phylum | NCBIreference | Similarity, %* | Pinus sylvestris | Picea abies | All | ||
|---|---|---|---|---|---|---|---|---|
| Roots | Peat | Roots | Peat | |||||
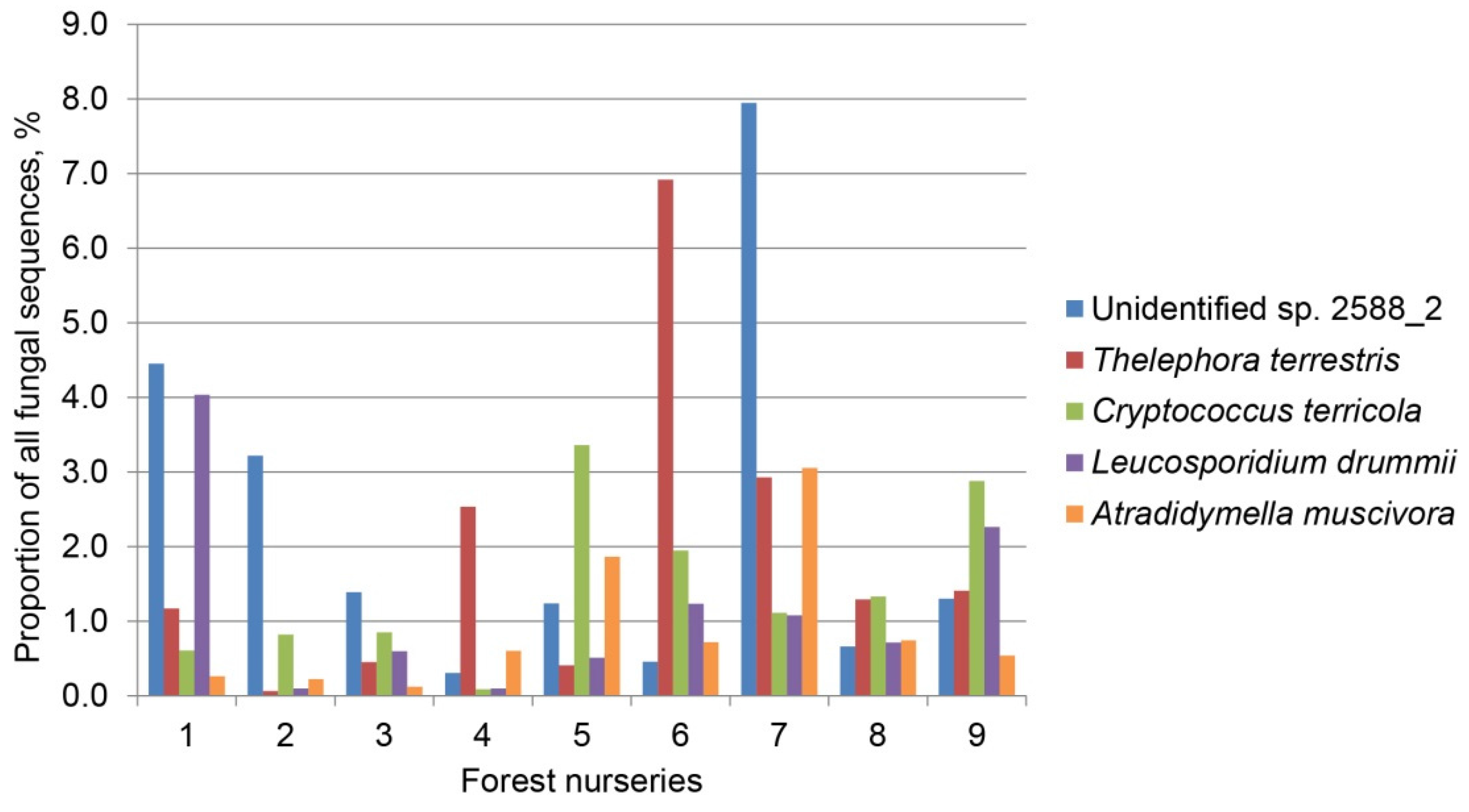
| Unidentified sp. 2588_2 | Ascomycota | JX317435 | 216/216 (100) | 41.6 | 9.1 | 43.1 | 16.5 | 21.0 |
| Thelephora terrestris | Basidiomycota | KP814380 | 329/331 (99) | 15.6 | 19.5 | 22.0 | 13.3 | 17.1 |
| Cryptococcus terricola | Basidiomycota | HF558655 | 342/347 (99) | 0.7 | 15.6 | 0.3 | 20.4 | 13.0 |
| Leucosporidium drummii | Basidiomycota | FN908919 | 323/324 (99) | 0.9 | 15.4 | 0.5 | 13.3 | 10.6 |
| Atradidymella muscivora | Ascomycota | EU817828 | 266/267 (99) | 8.1 | 7.2 | 7.5 | 9.4 | 8.1 |
| Phialophora sp. 2588_10 | Ascomycota | DQ069046 | 246/247 (99) | 5.3 | 2.1 | 1.6 | 2.8 | 2.8 |
| Umbelopsis isabellina | Mucoromycotina | EU484210 | 308/309 (99) | 0.1 | 2.4 | 0.2 | 3.7 | 2.2 |
| Trichoderma viride | Ascomycota | KT876533 | 199/200 (99) | 4.0 | 1.1 | 4.7 | 1.3 | 2.1 |
| Mortierella turficola | Mucoromycotina | JX976040 | 348/349 (99) | 0.4 | 4.4 | 0.0 | 0.6 | 1.9 |
| Laccaria proxima | Basidiomycota | KM067833 | 199/199 (100) | 0.8 | 2.2 | 0.9 | 2.1 | 1.8 |
| Rhodotorula fujisanensis | Basidiomycota | HF558662 | 327/328 (99) | 0.3 | 3.7 | 0.0 | 0.1 | 1.5 |
| Mortierella parvispora | Mortierellomycotina | JX976005 | 345/347 (99) | 0.2 | 1.8 | 0.3 | 1.9 | 1.4 |
| Oidiodendron rhodogenum | Ascomycota | NR_119425 | 252/252 (100) | 0.3 | 2.0 | 0.1 | 1.4 | 1.3 |
| Botrytis cinerea | Ascomycota | KR055052 | 206/206 (100) | 2.3 | 0.4 | 1.7 | 0.2 | 0.8 |
| Cladosporium cladosporioides | Ascomycota | KT151593 | 260/262 (99) | 2.3 | 0.6 | 0.6 | 0.4 | 0.8 |
| Meliniomyces variabilis | Ascomycota | KP753330 | 258/259 (99) | 0.8 | 0.0 | 4.5 | 0.2 | 0.7 |
| Tylospora fibrillosa | Basidiomycota | KR019831 | 306/306 (100) | - | - | 2.3 | 1.0 | 0.6 |
| Inocybe jacobi | Basidiomycota | HQ604812 | 290/292 (99) | 0.2 | 0.4 | 0.0 | 0.6 | 0.4 |
| Tylospora asterophora | Basidiomycota | HM190017 | 305/306 (99) | 0.0 | 0.0 | 2.1 | 0.3 | 0.4 |
| Cryptococcus sp. 2588_18 | Basidiomycota | KT372800 | 337/338 (99) | 0.1 | 0.6 | 0.0 | 0.2 | 0.3 |
| Suillus variegatus | Basidiomycota | JQ753773 | 276/277 (99) | 2.1 | 0.0 | - | 0.0 | 0.3 |
| Rhodotorula colostri | Basidiomycota | JX188225 | 317/319 (99) | 0.1 | 0.7 | 0.0 | 0.0 | 0.3 |
| Cystofilobasidium capitatum | Basidiomycota | HF558659 | 337/339 (99) | 0.0 | 0.8 | - | 0.0 | 0.3 |
| Fusarium avenaceum | Ascomycota | KT823772 | 275/276 (99) | 0.5 | 0.3 | 0.4 | 0.1 | 0.3 |
| Wilcoxina mikolae | Ascomycota | LC029799 | 273/274 (99) | 1.6 | 0.0 | 0.0 | 0.0 | 0.3 |
| Cryptococcus victoriae | Basidiomycota | KP299253 | 251/252 (99) | 0.3 | 0.4 | 0.0 | 0.1 | 0.3 |
| Unidentified sp. 2588_22 | Ascomycota | AM292201 | 274/274 (100) | 0.0 | 0.2 | 0.0 | 0.5 | 0.2 |
| Pezoloma ericae | Ascomycota | DQ069026 | 247/249 (99) | 0.5 | 0.2 | 0.5 | 0.1 | 0.2 |
| Amphinema sp. 2588_23 | Basidiomycota | JN943911 | 285/289 (99) | - | 0.0 | 0.6 | 0.5 | 0.2 |
| Lecythophora sp. | Ascomycota | KJ542236 | 263/267 (99) | 0.0 | 0.3 | 0.0 | 0.4 | 0.2 |
| Mortierella elongata | Mortierellomycotina | KP772767 | 361/364 (99) | 0.1 | 0.4 | 0.0 | 0.2 | 0.2 |
| Hebeloma cavipes | Basidiomycota | EU887517 | 320/321 (99) | 1.3 | 0.0 | - | - | 0.2 |
| Trichosporon porosum | Basidiomycota | HG737348 | 275/276 (99) | 0.6 | 0.1 | 0.1 | 0.2 | 0.2 |
| Total of 33 taxa | 90.9 | 92.1 | 94.3 | 91.9 | 92.1 | |||
| Forest nursery | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| Pinus sylvestris | |||||||||
| pH | 3.5 ± 0.1 cd | 3.2 ±0.1 d | 3.7 ± 0.00 cd | 4.0 ± 0.1 bc | 3.9 ± 0.2 c | 4.5 ± 0.0 ab | 3.9 ± 0.1 c | 3.8 ± 0.2 c | 4.5 ± 0.1 a |
| N (mg/L) | 0.7 ± 0.1 a | 2.3 ± 0.9 a | 1.1 ± 0.1 a | 1.2 ± 0.1 a | 0.7 ± 0.3 a | 0.4 ± 0.1 a | 0.6 ± 0.1 a | 1.2 ± 0.5 a | 0.5 ± 0.0 a |
| P (mg/L) | 7.8 ± 2.9 a | 5.6 ± 1.4 a | 5.5 ± 0.4 a | 7.9 ± 2.6 a | 8.9 ± 1.1 a | 12.1 ± 0.2 a | 7.5 ± 0.1 a | 7.5 ± 2.1 a | 6.4 ± 0.1 a |
| K (mg/L) | 114.5 ± 2.5 a | 131.5 ± 10.5 a | 115.0 ± 19.0 a | 135.0 ± 18.0 a | 105.0 ± 3.0 a | 91.3 ± 6.6 a | 94.6 ± 1.8 a | 93.5 ± 12.5 a | 88.2 ± 4.4 a |
| Ca (mg/L) | 16.2 ± 1.1 ab | 14.3 ± 0.3 b | 17.0 ± 1.2 ab | 18.4 ± 0.6 ab | 23.5 ± 3.2 a | 16.5 ± 0.4 ab | 19.9 ± 0.4 ab | 16.2 ± 0.1 ab | 23.9 ± 3.2 a |
| Mg (mg/L) | 5.6 ± 0.3 b | 5.2 ± 1.1 b | 5.9 ± 0.3 b | 7.7 ± 1.2 ab | 9.7 ± 0.3 a | 5.9 ± 0.1 b | 7.7 ± 0.0 ab | 5.1 ± 0.0 b | 8.6 ± 0.9 ab |
| Cl (mg/L) | 30.8 ± 4.4 a | 23.5 ± 2.9 a | 17.6 ± 2.9 a | 17.6 ± 11.7 a | 17.6 ± 2.9 a | 20.5 ± 2.9 a | 41.1 ± 5.9 a | 22.0 ± 4.4 a | 29.3 ± 8.8 a |
| Picea abies | |||||||||
| pH | 3.5 ± 0.0 b | 3.6 ± 0.1 b | 3.4 ± 0.2 b | 3.5 ± 0.1 b | 3.7 ± 0.1 ab | 3.7 ± 0.1 ab | 3.8 ± 0.2 ab | 3.6 ± 0.1 b | 4.5 ± 0.3 a |
| N (mg/L) | 3.0 ± 0.9 a | 6.1 ± 2.9 a | 2.0 ± 0.9 a | 2.4 ± 0.7 a | 1.2 ± 0.1 a | 0.5 ± 0.0 a | 0.4 ± 0.0 a | 6.8 ± 4.7 a | 0.5 ± 0.0 a |
| P (mg/L) | 9.8 ± 0.6 a | 9.9 ± 3.6 a | 5.4 ± 1.0 a | 6.9 ± 1.5 a | 4.7 ± 0.6 a | 8.7 ± 1.9 a | 6.9 ± 0.5 a | 7.8 ± 0.0 a | 12.6 ± 1.1 a |
| K (mg/L) | 117.5 ± 2.5 a | 129.5 ± 26.5 a | 107.5 ± 7.5 a | 152.5 ± 18.5 a | 88.6 ± 6.0 a | 124.5 ± 0.5 a | 96.7 ± 2.1 a | 99.5 ± 25.5 a | 111.0 ± 4.0 a |
| Ca (mg/L) | 14.7 ± 1.9 a | 29.6 ± 9.1 a | 18.1 ± 4.2 a | 15.9 ± 0.4 a | 19.9 ± 0.9 a | 21.4 ± 5.2 a | 20.2 ± 0.1 a | 23.3 ± 0.8 a | 24.7 ± 3.1 a |
| Mg (mg/L) | 5.1 ± 0.5 a | 12.5 ± 4.9 a | 5.7 ± 1.3 a | 5.1 ± 0.1 a | 6.2 ± 0.2 a | 7.5 ± 1.7 a | 7.6 ± 0.2 a | 7.6 ± 0.1 a | 8.6 ± 0.2 a |
| Cl (mg/L) | 23.5 ± 2.9 ab | 23.5 ± 2.9 ab | 13.2 ± 1.5 b | 16.2 ± 1.5 ab | 24.9 ± 1.5 ab | 24.9 ± 4.4 ab | 41.0 ± 11.7 a | 17.6 ± 2.9 ab | 29.3 ± 8.8 ab |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menkis, A.; Burokienė, D.; Stenlid, J.; Stenström, E. High-Throughput Sequencing Shows High Fungal Diversity and Community Segregation in the Rhizospheres of Container-Grown Conifer Seedlings. Forests 2016, 7, 44. https://doi.org/10.3390/f7020044
Menkis A, Burokienė D, Stenlid J, Stenström E. High-Throughput Sequencing Shows High Fungal Diversity and Community Segregation in the Rhizospheres of Container-Grown Conifer Seedlings. Forests. 2016; 7(2):44. https://doi.org/10.3390/f7020044
Chicago/Turabian StyleMenkis, Audrius, Daiva Burokienė, Jan Stenlid, and Elna Stenström. 2016. "High-Throughput Sequencing Shows High Fungal Diversity and Community Segregation in the Rhizospheres of Container-Grown Conifer Seedlings" Forests 7, no. 2: 44. https://doi.org/10.3390/f7020044






