Flow Chamber System for the Statistical Evaluation of Bacterial Colonization on Materials
Abstract
:1. Introduction
2. Results
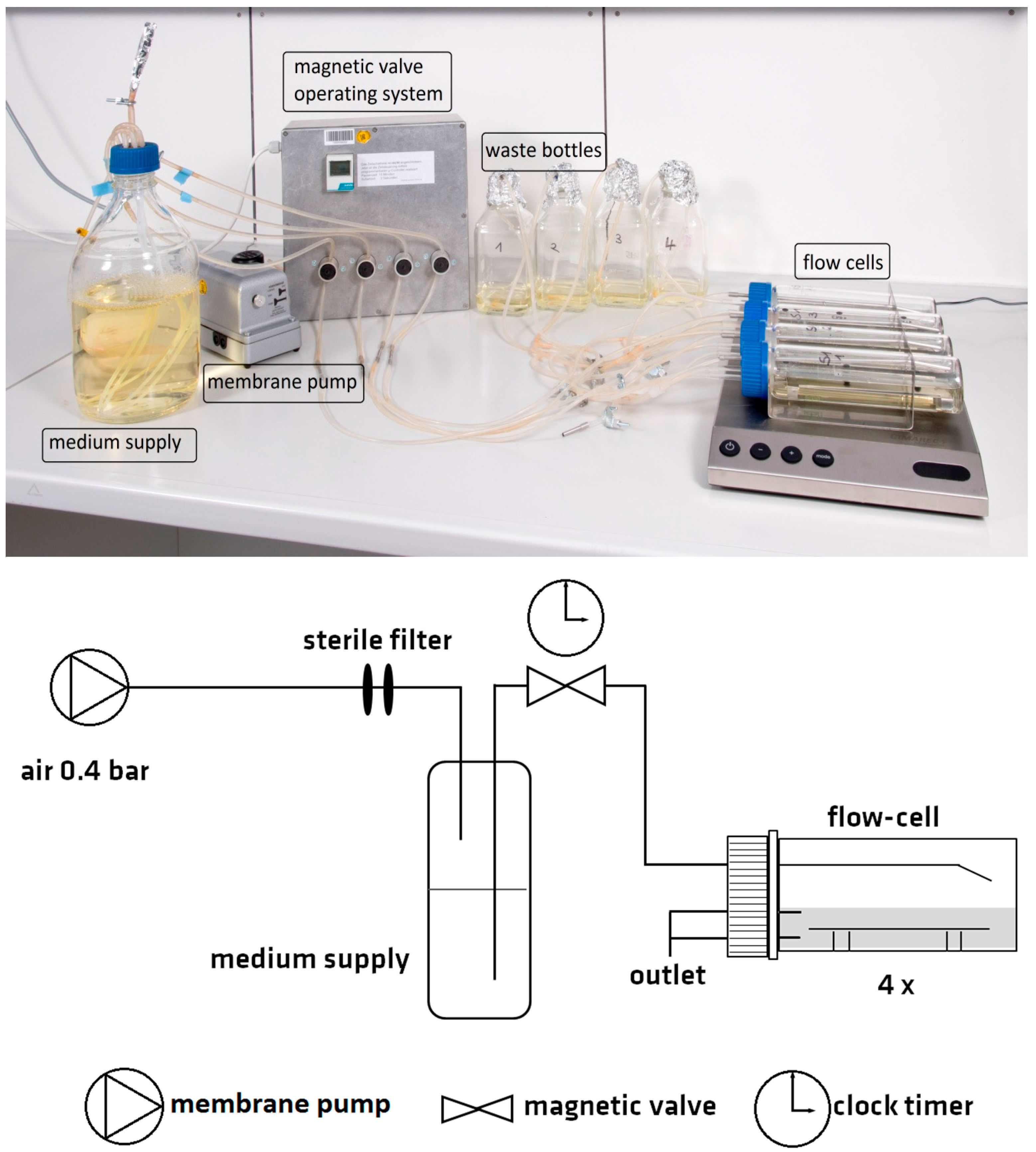
2.1. Design of the Flow Chamber System
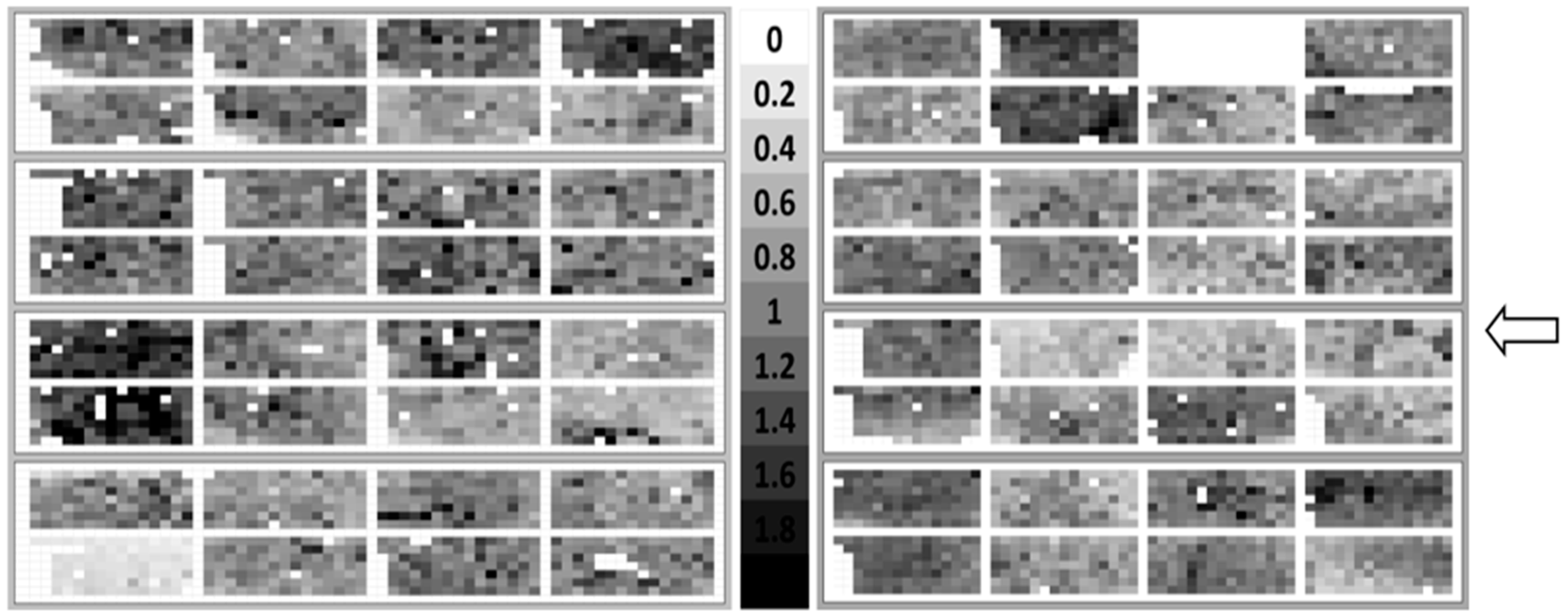
2.2. Quantification of the Initial Bacterial Adhesion
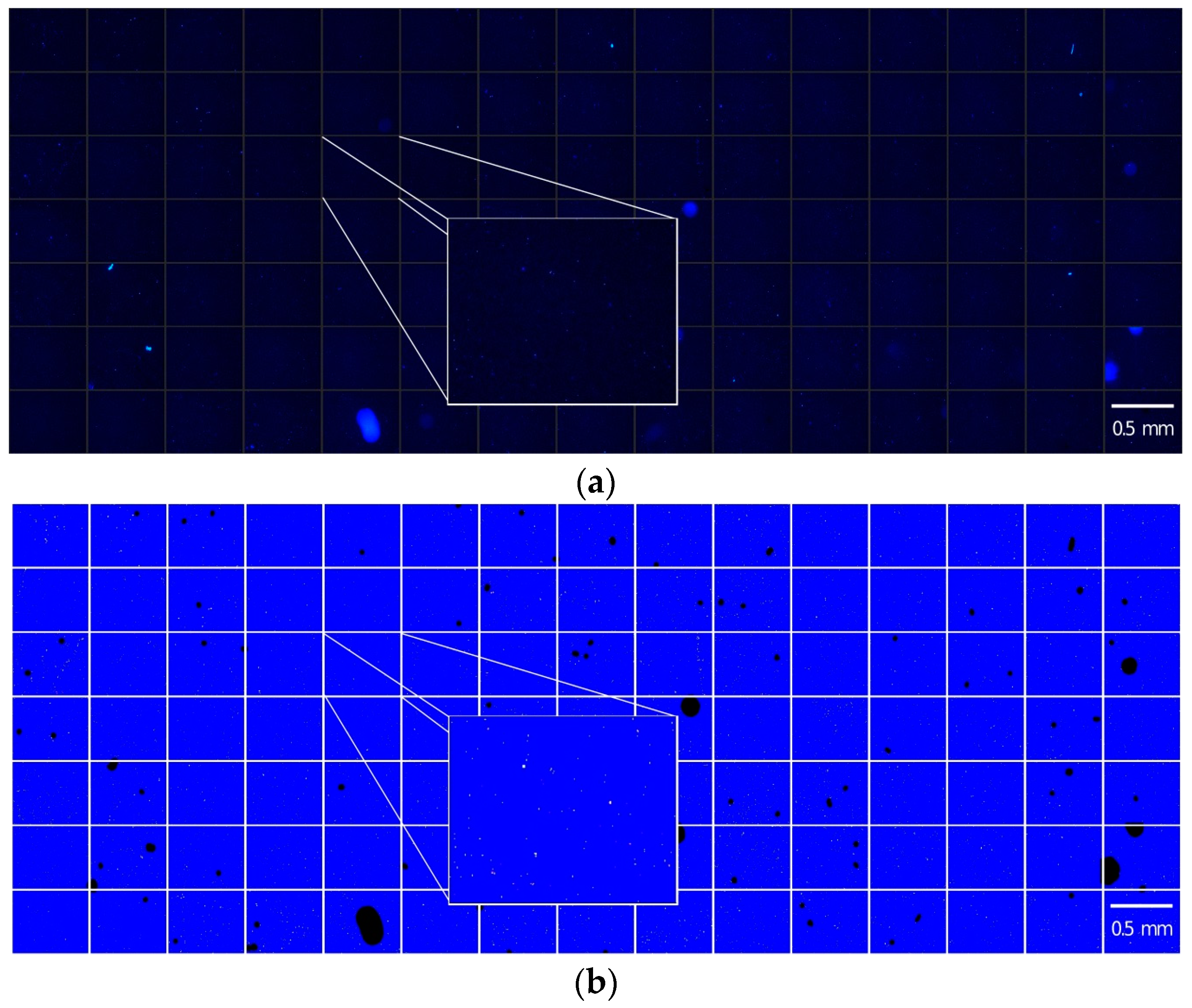
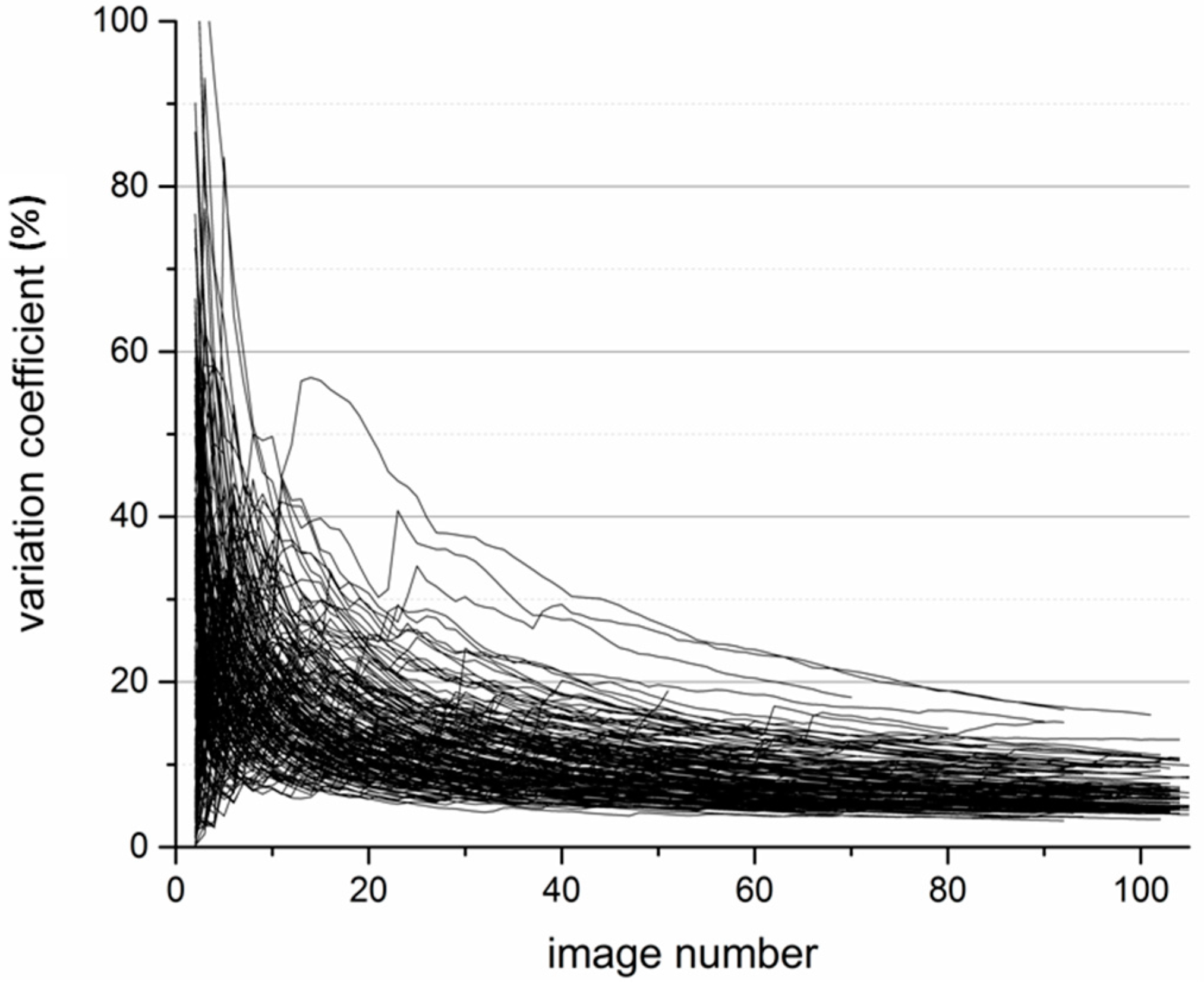
2.3. Determination of the Minimum Image Number
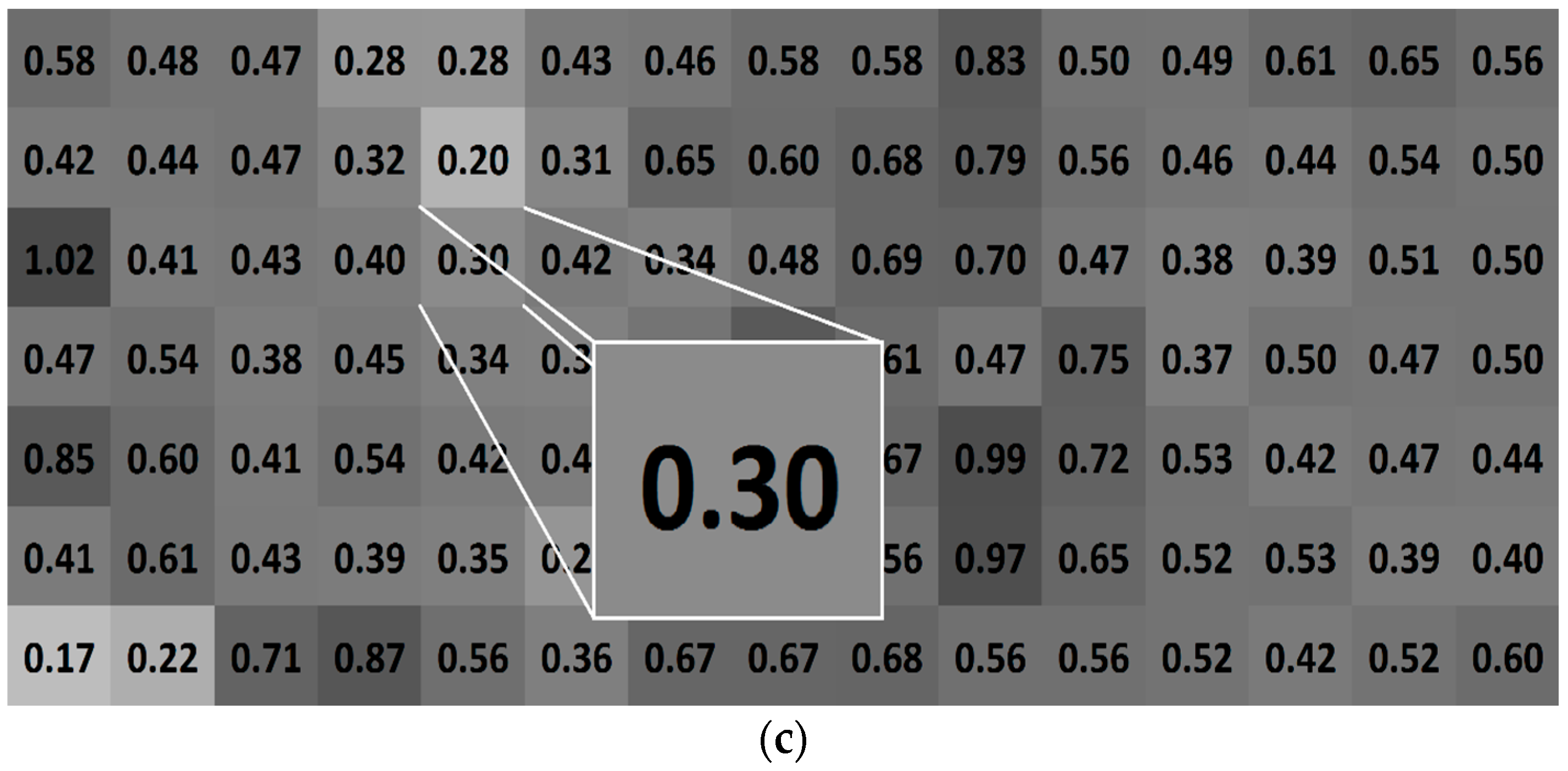
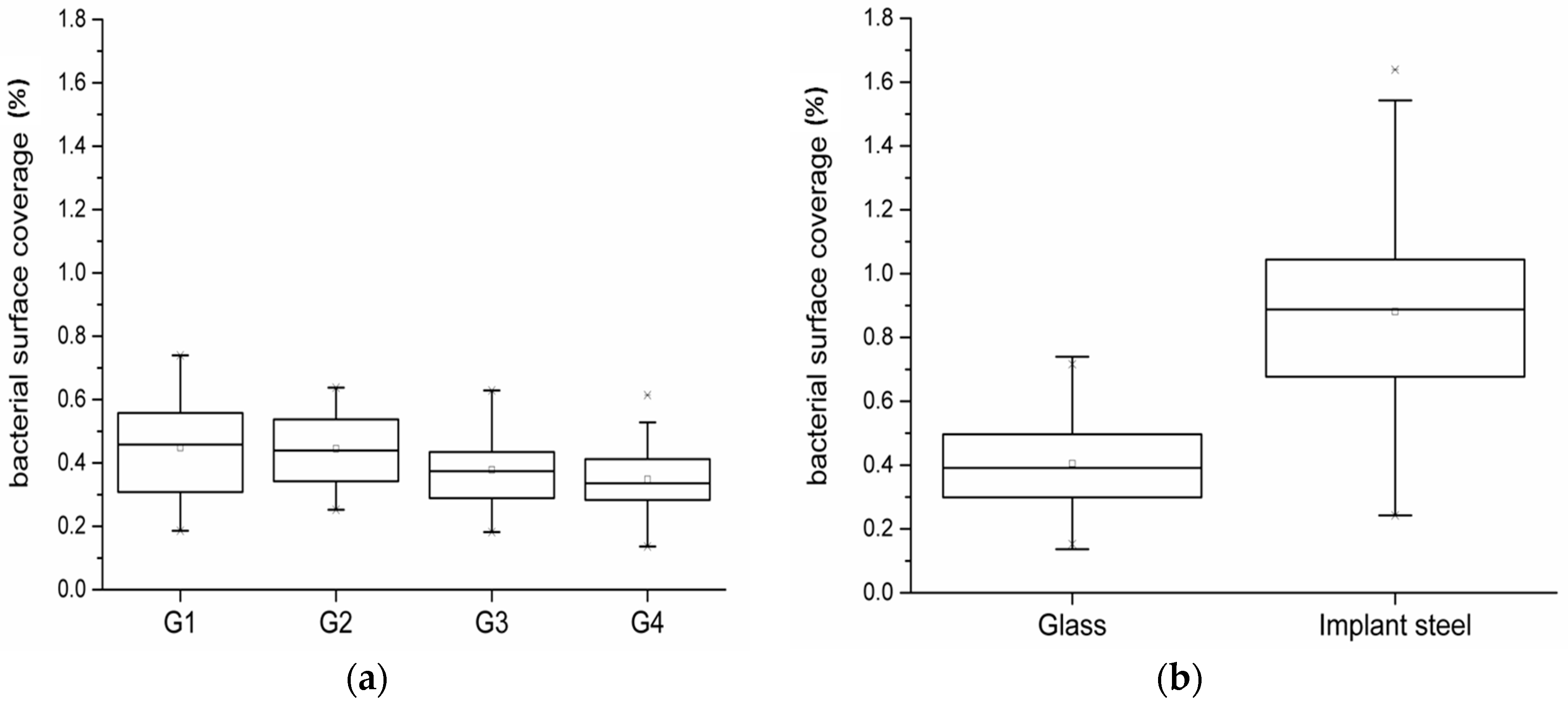
2.4. Assessment of Bacterial Adhesion in Dependence of the Coupon Position in the Flow Chamber System
2.5. Reproducibility of Results
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Growth Media
4.2. Test Coupons
4.3. Operation of the Flow Chamber System
4.4. Bacterial Attachment Tests
4.5. Microscopy
4.6. Image Analysis
4.7. Statistical Evaluation
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ANOVA | analysis of variance |
| CFU | colony forming units |
| DO | dissolved oxygen |
Appendix A. Evaluation of the Influence of the Flow Conditions on the Media Distribution within the Flow Chamber and the Colonization Pattern


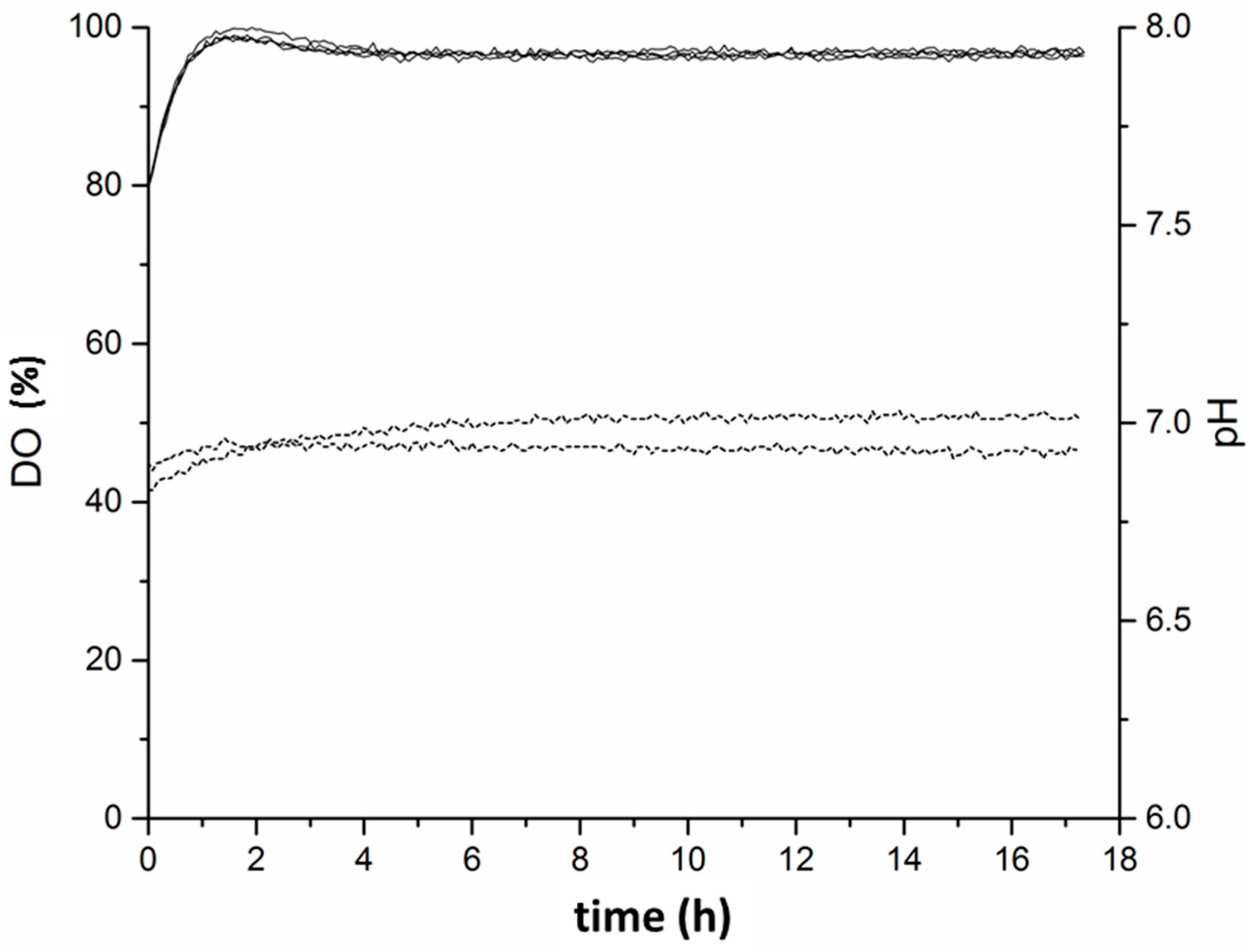
Appendix B. DO and pH Measurement
References
- Dufour, D.; Leung, V.; Levesque, C.M. Bacterial biofilm: Structure, function, and antimicrobial resistance. Endod. Top. 2012, 22, 2–16. [Google Scholar] [CrossRef]
- Garrett, T.R.; Bhakoo, M.; Zhang, Z.B. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- Schultz, M.P.; Bendick, J.A.; Holm, E.R.; Hertel, W.M. Economic impact of biofouling on a naval surface ship. Biofouling 2011, 27, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilm formation: A clinically relevant microbiological process. Clin. Infect. Dis. 2001, 33, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef] [PubMed]
- Valappil, S.P. Antimicrobial interfaces. In Biointerfaces: Where Material Meets Biology; Royal Society of Chemistry (RSC): London, UK, 2014; pp. 399–423. [Google Scholar]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.; Flint, S.; Brooks, J. Bacterial cell attachment, the beginning of a biofilm. J. Ind. Microbiol. Biotechnol. 2007, 34, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Cunha, A.; Elie, A.M.; Plawinski, L.; Serro, A.P.; Rego, A.M.B.; Almeida, A.; Urdaci, M.C.; Durrieu, M.C.; Vilar, R. Femto second laser surface texturing of titanium as a method to reduce the adhesion of Staphylococcus aureus and biofilm formation. Appl. Surf. Sci. 2016, 360, 485–493. [Google Scholar] [CrossRef]
- Javed, M.A.; Stoddart, P.R.; McArthur, S.L.; Wade, S.A. The effect of metal microstructure on the initial attachment of Escherichia coli to 1010 carbon steel. Biofouling 2013, 29, 939–952. [Google Scholar] [CrossRef] [PubMed]
- Genzer, J.; Efimenko, K. Recent developments in superhydrophobic surfaces and their relevance to marine fouling: A review. Biofouling 2006, 22, 339–360. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.F. On the nature of biomaterials. Biomaterials 2009, 30, 5897–5909. [Google Scholar] [CrossRef] [PubMed]
- Pantanella, F.; Valenti, P.; Natalizi, T.; Passeri, D.; Berlutti, F. Analytical techniques to study microbial biofilm on abiotic surfaces: Pros and Cons of the main techniques currently in use. Ann. Ig. 2013, 25, 31–42. [Google Scholar] [PubMed]
- Zaborowska, M.; Welch, K.; Branemark, R.; Khalilpour, P.; Engqvist, H.; Thomsen, P.; Trobos, M. Bacteria-material surface interactions: Methodological development for the assessment of implant surface induced antibacterial effects. J. Biomed. Mater. Res. B Appl. Biomater. 2015, 103, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.I.; Sydnes, L.K.; Landfald, B.; Strom, A.R. Osmoregulation in Escherichia coli by accumulation of organic osmolytes: Betaines, glutamic acid, and trehalose. Arch. Microbiol. 1987, 147, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zack, G.W.; Rogers, W.E.; Latt, S.A. Automatic measurement of sister chromatid exchange frequency. J. Histochem. Cytochem. 1977, 25, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Ludecke, C.; Jandt, K.D.; Siegismund, D.; Kujau, M.J.; Zang, E.; Rettenmayr, M.; Bossert, J.; Roth, M. Reproducible biofilm cultivation of chemostat-grown Escherichia coli and investigation of bacterial adhesion on biomaterials using a non-constant-depth film fermenter. PLoS ONE 2014, 9, e84837. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Nelis, H.J. In vitro and in vivo model systems to study microbial biofilm formation. J. Microbiol. Methods 2010, 83, 89–105. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.V.; Mosier, A.P.; Kiehl, T.R.; Kaloyeros, A.E.; Cady, N.C. Development of antifouling surfaces to reduce bacterial attachment. Soft Matter 2013, 9, 6235–6244. [Google Scholar] [CrossRef]
- Gillett, A.; Waugh, D.; Lawrence, J.; Swainson, M.; Dixon, R. Laser surface modification for the prevention of biofouling by infection causing Escherichia Coli. J. Laser Appl. 2016, 28, 022503. [Google Scholar] [CrossRef]
- Buckingham-Meyer, K.; Goeres, D.M.; Hamilton, M.A. Comparative evaluation of biofilm disinfectant efficacy tests. J. Microbiol. Methods 2007, 70, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.; Zhou, Y.; Pinkner, J.S.; Dodson, K.W.; Crowley, J.R.; Heuser, J.; Chapman, M.R.; Hadjifrangiskou, M.; Henderson, J.P.; Hultgren, S.J. Escherichia coli biofilms have an organized and complex extracellular matrix structure. MBio 2013, 4, e00645-13. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Reisner, A.; Krogfelt, K.A.; Klein, B.M.; Zechner, E.L.; Molin, S. In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: Impact of environmental and genetic factors. J. Bacteriol. 2006, 188, 3572–3581. [Google Scholar] [CrossRef] [PubMed]
- Dewanti, R.; Wong, A.C. Influence of culture conditions on biofilm formation by Escherichia coli O157:H7. Int. J. Food Microbiol. 1995, 26, 147–164. [Google Scholar] [CrossRef]
- D’Souza, F.; Bruin, A.; Biersteker, R.; Donnelly, G.; Klijnstra, J.; Rentrop, C.; Willemsen, P. Bacterial assay for the rapid assessment of antifouling and fouling release properties of coatings and materials. J. Ind. Microbiol. Biot. 2010, 37, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Oethinger, M.; Tuohy, M.J.; Procop, G.W.; Bauer, T.W. Improved detection of biofilm-formative bacteria by vortexing and sonication: A pilot study. Clin. Orthop. Relat. Res. 2009, 467, 1360–1364. [Google Scholar] [CrossRef] [PubMed]
- Bonkat, G.; Rieken, M.; Rentsch, C.A.; Wyler, S.; Feike, A.; Schafer, J.; Gasser, T.; Trampuz, A.; Bachmann, A.; Widmer, A.F. Improved detection of microbial ureteral stent colonisation by sonication. World J. Urol. 2011, 29, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Van de Belt, H.; Neut, D.; Schenk, W.; van Horn, J.R.; van der Mei, H.C.; Busscher, H.J. Staphylococcus aureus biofilm formation on different gentamicin-loaded polymethylmethacrylate bone cements. Biomaterials 2001, 22, 1607–1611. [Google Scholar] [CrossRef]
- Nguyen, L.L.; Nelson, C.L.; Saccente, M.; Smeltzer, M.S.; Wassell, D.L.; McLaren, S.G. Detecting bacterial colonization of implanted orthopaedic devices by ultrasonication. Clin. Orthop. Relat. Res. 2002, 403, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Trampuz, A.; Piper, K.E.; Jacobson, M.J.; Hanssen, A.D.; Unni, K.K.; Osmon, D.R.; Mandrekar, J.N.; Cockerill, F.R.; Steckelberg, J.M.; Greenleaf, J.F.; et al. Sonication of removed hip and knee prostheses for diagnosis of infection. N. Engl. J. Med. 2007, 357, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Zacheus, O.M.; Iivanainen, E.K.; Nissinen, T.K.; Lehtola, M.J.; Martikainen, P.J. Bacterial biofilm formation on polyvinyl chloride, polyethylene and stainless steel exposed to ozonated water. Water Res. 2000, 34, 63–70. [Google Scholar] [CrossRef]
- Schmidt-Emrich, S.; Stiefel, P.; Rupper, P.; Katzenmeier, H.; Amberg, C.; Maniura-Weber, K.; Ren, Q. Rapid Assay to Assess Bacterial Adhesion on Textiles. Materials 2016, 9, 249. [Google Scholar] [CrossRef]
- Buchholz, F.; Lerchner, J.; Mariana, F.; Kuhlicke, U.; Neu, T.R.; Harms, H.; Maskow, T. Chip-calorimetry provides real time insights into the inactivation of biofilms by predatory bacteria. Biofouling 2012, 28, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Nam, J.H.; Kim, Y.H.; Lee, K.H.; Lee, D.H. Bacterial communities in the initial stage of marine biofilm formation on artificial surfaces. J. Microbiol. 2008, 46, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Sandt, C.; Smith-Palmer, T.; Pink, J.; Brennan, L.; Pink, D. Confocal Raman microspectroscopy as a tool for studying the chemical heterogeneities of biofilms in situ. J. Appl. Microbiol. 2007, 103, 1808–1820. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menzel, F.; Conradi, B.; Rodenacker, K.; Gorbushina, A.A.; Schwibbert, K. Flow Chamber System for the Statistical Evaluation of Bacterial Colonization on Materials. Materials 2016, 9, 770. https://doi.org/10.3390/ma9090770
Menzel F, Conradi B, Rodenacker K, Gorbushina AA, Schwibbert K. Flow Chamber System for the Statistical Evaluation of Bacterial Colonization on Materials. Materials. 2016; 9(9):770. https://doi.org/10.3390/ma9090770
Chicago/Turabian StyleMenzel, Friederike, Bianca Conradi, Karsten Rodenacker, Anna A. Gorbushina, and Karin Schwibbert. 2016. "Flow Chamber System for the Statistical Evaluation of Bacterial Colonization on Materials" Materials 9, no. 9: 770. https://doi.org/10.3390/ma9090770
APA StyleMenzel, F., Conradi, B., Rodenacker, K., Gorbushina, A. A., & Schwibbert, K. (2016). Flow Chamber System for the Statistical Evaluation of Bacterial Colonization on Materials. Materials, 9(9), 770. https://doi.org/10.3390/ma9090770




