Microarray Analysis of Mercury-Induced Changes in Gene Expression in Human Liver Carcinoma (HepG2) Cells: Importance in Immune Responses
Abstract
:Introduction
Materials and Methods
Chemical Reagents and Growth medium
Cell Culture and Harvesting
Mercury Exposure
Total RNA Extraction, Stabilization and Quantitation
Hybridization of Biotin-labeled cRNA to Oligonucleotide Microarrays and Staining Procedures
Microarray Image and Probe Array Scanning
Statistical Analysis
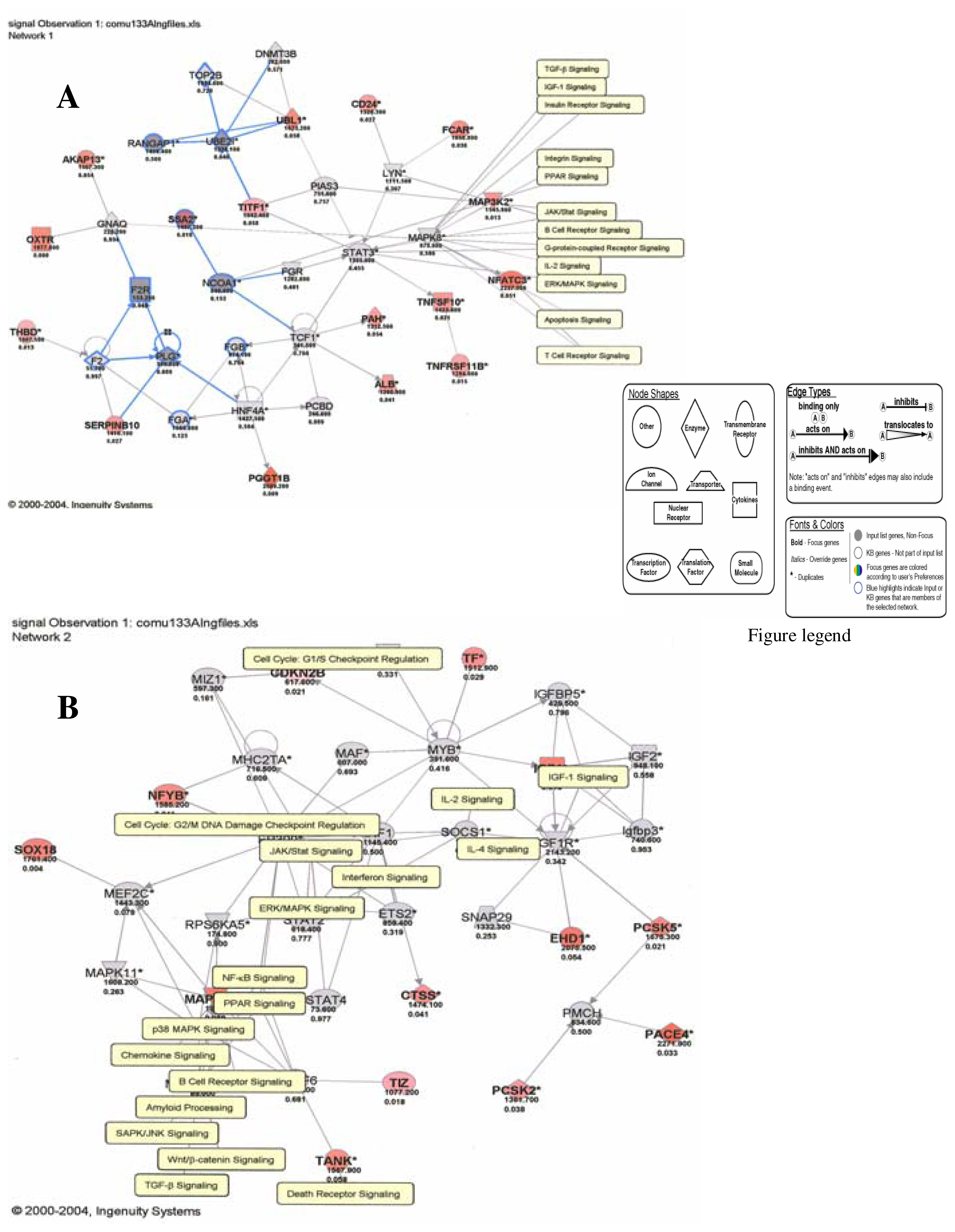
Results
Discussion
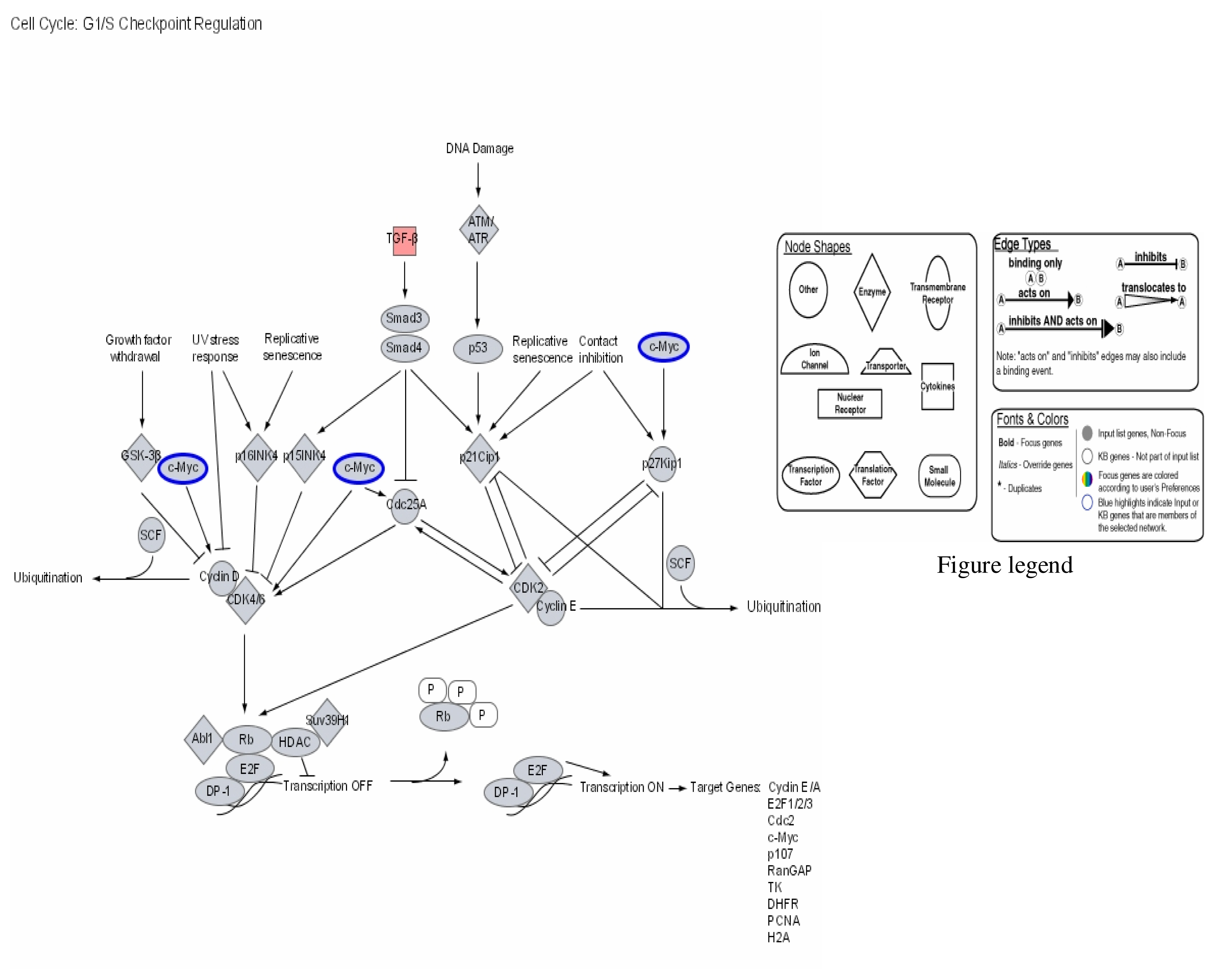
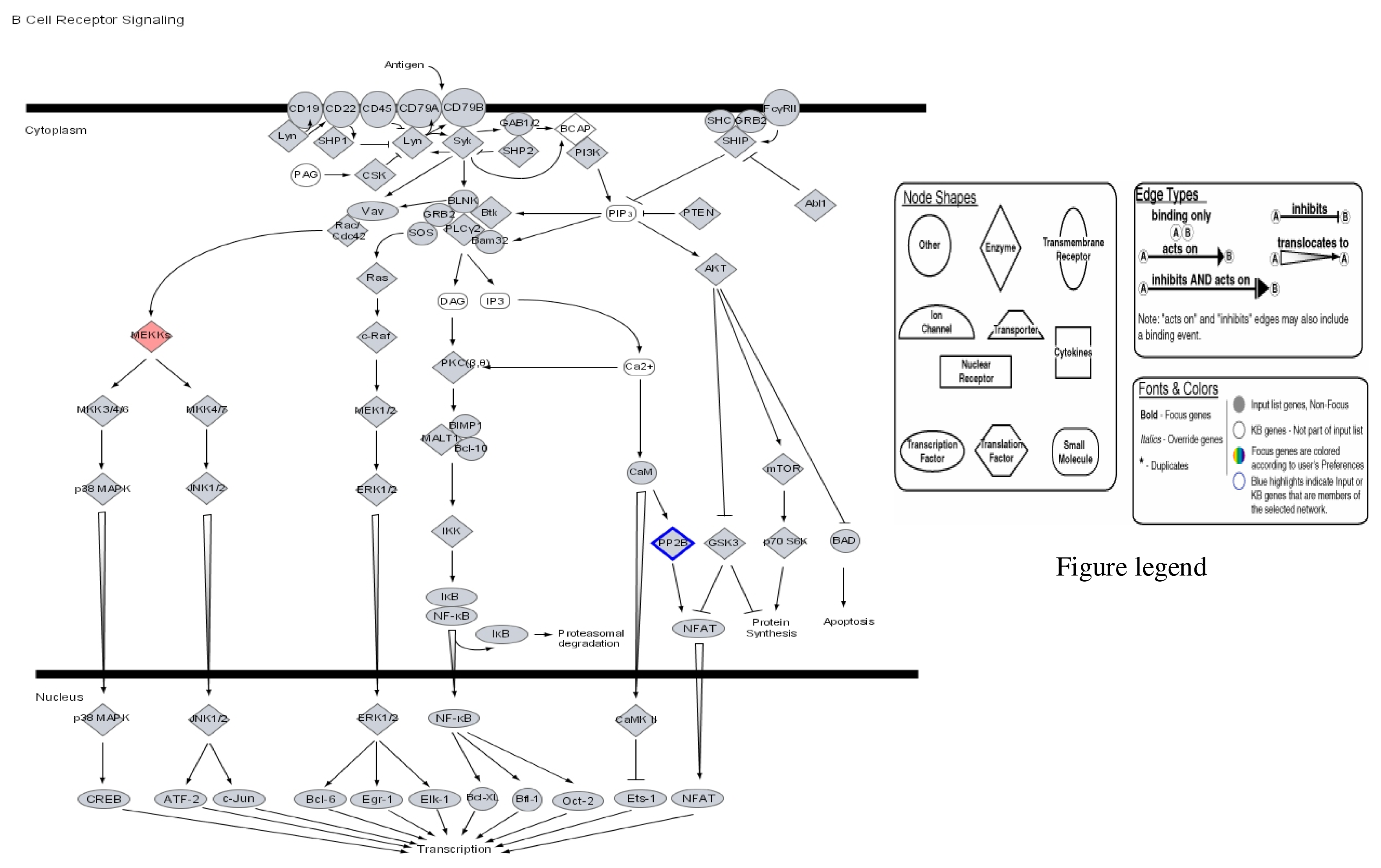
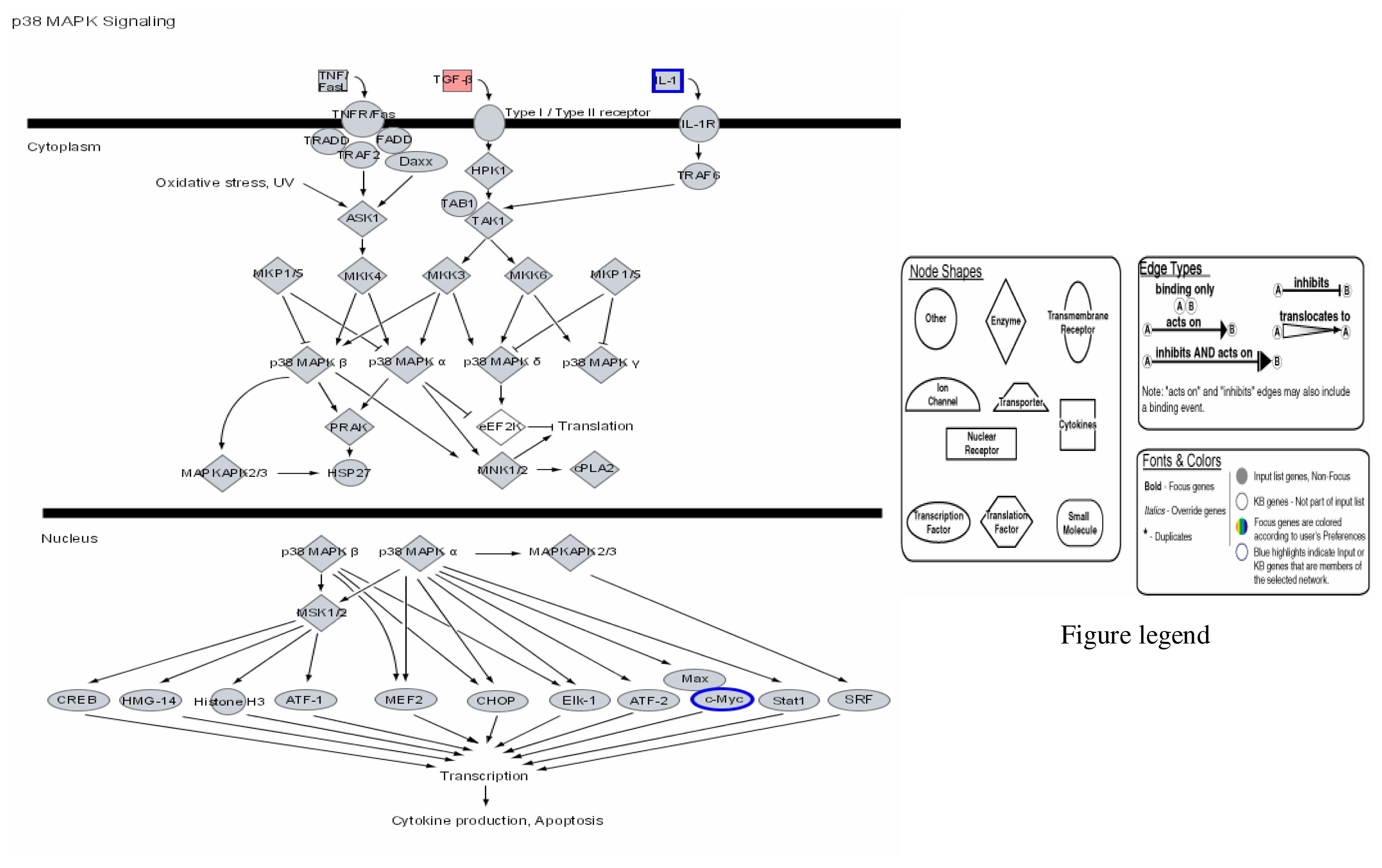
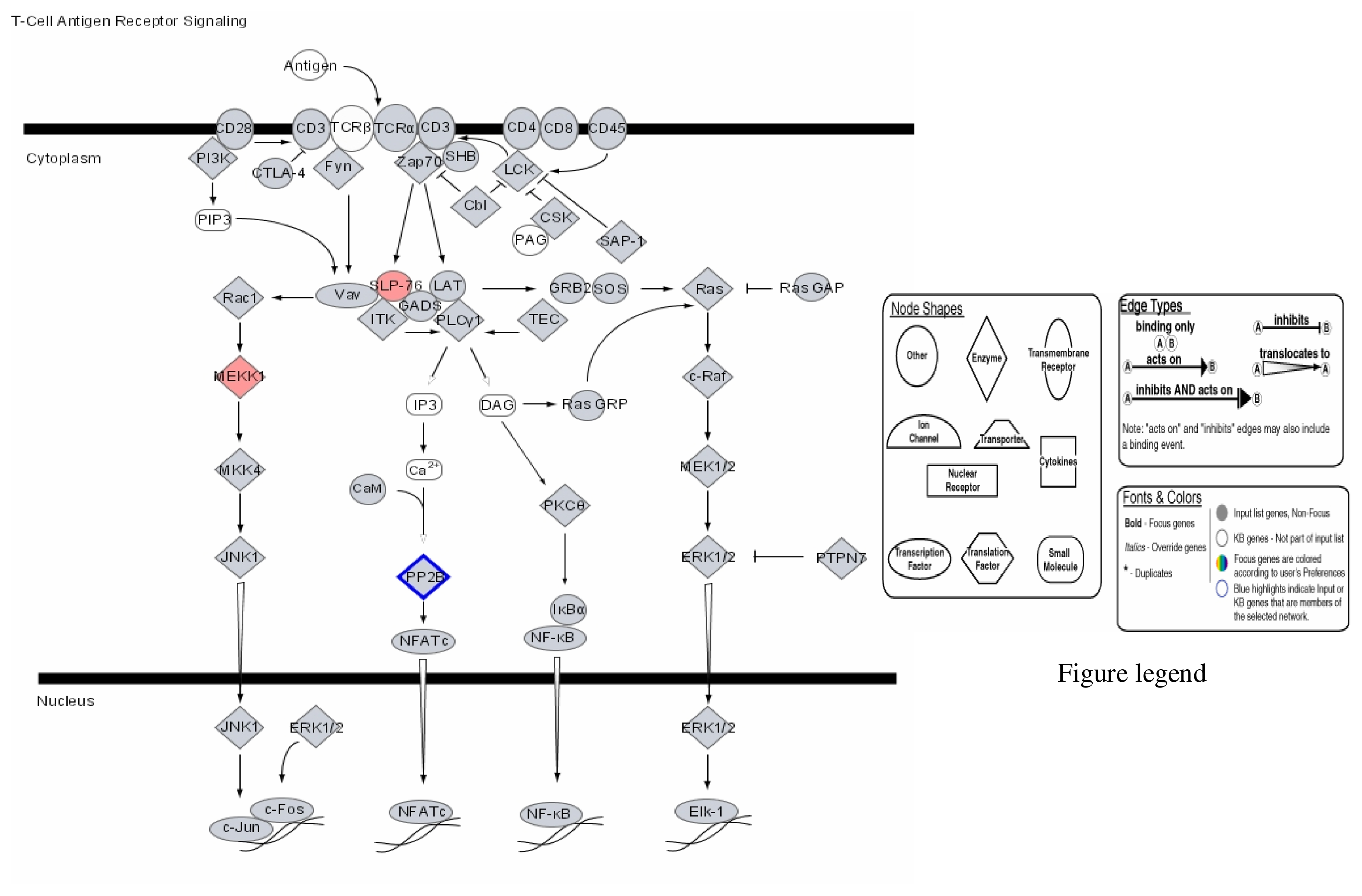
| Probe Set ID (Affymetrix) | Gene Title (Target Description/Symbol) | Chromosome Location | 1μg/mL Hg SLR Change/Fold Change | 2μg/mL Hg SLR Change/Fold Change | 3μg/mL Hg SLR Change/Fold Change |
|---|---|---|---|---|---|
| 209908_s_at | Transcript assignment(s)-GENSCAN0000002140 cdna: Genscan chromosome: NCBI35: 1:214900212:214908104:1 Alignment(s): Chr 1: 214907888–215003873(+) UCSC. Protein similarities: blast P09858-Transforming growth factor beta 2 precursor (TGF-beta 2); E value 0.0 Pfam-IPR001111 EMBL.EBI-Transforming growth factor beta (TGFb), N-terminal Pfam-IPR001839 EMBL.EBI -Transforming growth factor beta. | Align chr 1 | 5/32 | 4/16 | 1/2 |
| 206814_at | Nerve growth factor, beta polypeptide nerve growth factor, signal transducer activity/NGFB HGNC/—Cell-cell signaling development; 6112 energy reserve metabolism; 7166 cell surface receptor linked signal transduction; 7275 development; 4896 hematopoietin/interferon-class (D200-domain) cytokine receptor activity. | 1p13.1 | 1/2 | 2/4 | 3/8 |
| 208498_s_at | Amylase, alpha 1A; 1B; 1C (salivary); Amylase 2A, 2B(Pancreatic); AMY1A; AMY1B; AMY1C HGNC: AMY2A: AMY2B HGNC; 5975 carbohydrate metabolism; 7586 digestion; 5615 Extracellular space; 4556 alpha amylase activity; 5509 Calcium ion binding; 16798 hydrolase activity, acting on glycosyl bonds; 4556 alpha amylase activity; Pathway: starch and sucrose metabolism GENMAPP/KEGG. | 1p21 | 5/32 | 7/128 | 4/16 |
| 211096_at | pre-B-cell leukemia transcription factor 2; PBX2 HGNC; 6355 regulation of transcription; 7387 anterior compartment specification; 7388 posterior compartment specification; 15977 carbon utilization by fixation of carbon dioxide; 5634 nucleus; 9573 ribulose bisphosphate carboxylase complex; 3700 transcription factor activity; 16984 ribulose-bisphosphate carboxylase activity. | 1p21.3 | 5/32 | 4/16 | 0/1 |
| 213496_at | Plasticity related gene 1/LPPR4 HGNC; brain-specific phosphatidic acid phosphatase-like protein 1 [homo sapiens] | 1p21.3 | 5/32 | 4/16 | 0/1 |
| 207255_at | Leptin receptor, LEPR HGNC; cytokine receptor gp130 cytokine binding-domains. | 1p31 | 0/1 | −1/−2 | −1/−2 |
| 211703_s_at | Precursor beta-amyloid binding protein; BBP HGNC. | 1p32.1 | 2/4 | 4/16 | 4/16 |
| 220611_at | Disabled homologue 1 (Drosophila); Development, neurogenesis; phosphotyrosine interaction domain. See above. | 1p32-p31 | 2/4 | 4/16 | 4/16 |
| 205323_s_at | Metal-regulatory transcription factor 1; MTF1 HGNC; regulation of transcription from Pol 11 promoter; response to metal ion; transcription factor activity; transcription co-activator activity; zinc ion binding. | 1p33 | 2/4 | 1/2 | 2/4 |
| 208628_s_at | Nuclease sensitive element binding protein 1; NSEP1 HGNC; 6355 regulation of transcription, DNA dependent; 6366 transcription from Pol 11 promoter; 9613 response to pest/pathogen/parasite; 5634 nucleus; 3677 DNA binding; 3690 double-stranded DNA binding; 3697 single-stranded DNA binding; 3700 transcription factor activity; 16564 transcriptional repressor activity. | 1p34 | 3/8 | 1/2 | 1/2 |
| 219925_at | Zinc finger protein 258;ZN258 HGNC; development; membrane; DNA binding; | 1p34.2 | 3/8 | 1/2 | 1/2 |
| 215268_at | KIAA 0754 protein; KIAA0754 HGNC; Protein similarities: blast ID XP 513340-Predicted: microfilament and actin filament cross-linker protein (Pan troglodytes); E value 0.0; blast ID BAC87042.1 unnamed protein product (Homo sapiens). | 1p34.3 | 8/256 | 8/256 | 6/64 |
| 203503_s_at | Peroxisomal biogenesis factor 14; PEX14 HGNC/peroxisomal anchor protein (Fragment); 5777 peroxisome; 5779 integral to peroxixomal membrane; 16020 membrane. | 1p36.22 | 3/8 | 2/4 | 2/4 |
| 218731_s_at | von Willebrand factor A domain-related protein; WARP HGNC. | 1p36.33 | 3/8 | 2/4 | 3/8 |
| 200006_at | 7 Parkinson disease (autosomal recessive, early onset); PARK7 HGNC; Overlapping Transcripts: NM_007262 NCBI chromosome1: 7731338–7754882 (+) UCSC. | 1p36.33-p36.12 | 3/8 | 2/4 | 3/8 |
| 214130_s_at | Phosphodiesterase 4D interacting protein (myomegadin); PDE4DIP HGNC; 6412 protein biosynthesis; 7010 cytoskeleton organization and biogenesis; 30036 actin cytoskeleton organization and biogenesis; 5622 intracellular; 5840 ribosome; 15629 actin cytoskeleton; 3735 structural constituent of ribosome; 3779 actin binding. | 1q12 | 5/32 | 5/32 | 1/2 |
| 205076_s_at | Cisplatin resistance associated CRA HGNC. | 1q12-q21 | −1/−2 | −1/−2 | −1/−2 |
| 217048_at | SHC (Src homology 2 domain containing) transforming protein 1 SHC1 HGNC; 187 activation of MAPK; 1558 regulation of cell growth; 7176 regulation of epidermal growth factor receptor activity; 7242 intracellular signaling cascade; 8284 positive regulation of cell proliferation; 45840 positive regulation of mitosis; 5886 plasma membrane; 5068 transmembrane receptor protein tyrosine kinase adaptor protein activity. 5543 phospholipid binding; Pathway-Integrin-mediated_cell_adhesion GENMAPP/KEGG. | 1q21 | −1/−2 | −1/−2 | −1/−2 |
| 203411_s_at | Lamin A/C, Intermediate filament protein; muscle development. | 1q21.2-q21.3 | −6/−64 | −4/−16 | −4/−16 |
| 200652_at | Signal sequence receptor, beta (transloxon-associated protein beta); SSR2 HGNC; 6613 cotranslational membrane targeting; 5783 endoplasmic reticulum; 16021 integral membrane; 5048 signal sequence binding; | 1q21-q23 | −6/−64 | −4/−16 | −4/−16 |
| 204007_at | Fc fragment of IgG, low affinity llla, receptor for (CD16)/FCGR3A HGNC; low affinity immunoglobulin γ Fc region receptor III-B precursor (IgG Fc receptor III-1) Fc-γ RIII-β) Fc-γ RIIIB), (Fc-γRIII), (FcRIII) (CD16-B)(FcR-10). | 1q23 | −2/−4 | −1/−2 | −2/−4 |
| 201966_at | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase); NDUFS2 HGNC; 6118 Electron transport; 6120 mitochondrial electron transport, NADH to ubiquionone; 56243 membrane fraction; 5739 mitochondrion; 3954 NADH dehaydrogenase) activity; 8137 NADH dehaydrogenase activity (ubiquinone); 9055 electron carrier activity; 16491 oxidoreductase activity; 5489 complex 1_49Kd; electron transporter activity; 1.4e-205; Pathway: Electron_Transport_Chain GENMAPP/KEGG. | 1q23 | −2/−4 | −1/−2 | 0/1 |
| 235401_s_at | Fc receptor homolog expressed in B cells; FREB HGNC; 4872-receptor activity. | 1q23.1 | −2/−4 | −1/−2 | −2/−4 |
| 218229_s_at | Pogo transposable element with KRAB domain/POGK HGNC; regulation of transcription, DNA-dependent; anti-apoptosis; development. | 1q23.3 | −2/4 | −1/2 | −3/−8 |
| 211188_at | CD84 antigen (leucocyte antigen); CD84 HGNC; 6952 defence response; 7156 homophilic cell adhesion; 5887 integral to plasma membrane. | 1q24 | −5/−32 | −6/−64 | −4/−16 |
| 200844_s_at | Peroxiredoxin 6 PRDX6 HGNC;/antioxidant protein 2; non-selenium glutathione peroxidase. | 1q24.2 | −5/−32 | −6/−64 | −4/−16 |
| 206245_s_at | Influenza virus NS1A binding protein; IVNS1ABP HGNC; 6383 transcription from Pol lll promoter; 8380RNA splicing; 9615 response to virus; 5667 transcription factor complex; 5681 spliceosome complex; 5515 protein binding. | 1q25.1-q31.1 | 0/1 | −1/−2 | −3/−8 |
| 217602_at | Ubiquitin specific protease 39/USP39 HGNC; ubiquitin carboxy-terminal hydrolase family protein, ubiquitin protease, spliceosome assembly; mRNA processing; RNA splicing, cysteine-type endopeptidase activity; pre-mRNA splicing factor activity. | 2p11.2 | 5/32 | 3/8 | 2/2 |
| 205871_at | Plasminogen-like; PLGL HGNC; plasmin activity not recorded. | 2p11-q11 | 5/32 | 3/8 | 5/32 |
| 200671_s_at | Spectrin, beta, non-erythrocytic1/SPTBN1 HGNC; Duchenne muscular dystrophy protein, brain isoform (fragment); beta spectrin 2. | 2p21 | 4/16 | 3/8 | 2/4 |
| 211615_s_at | Leucine-rich PPR-motif containing/LRPPRC HGNC; bicoid stability factor. | 2p22.1 | 0/1 | −2−4 | 0/1 |
| 219020_at | HS1-binding protein 3 /FLJ14249 HGNC; intracellular signaling cascade | 2p24.2 | 0/1 | −1/−2 | −1/−2/ |
| 218495_at | Ribonuclease H1; RNASEH1 HGNC/ magnesium ion binding, RNA binding, endonuclease activity, ribonuclease H activity, hydrolase activity, maseH; nucleic acid binging | 2p25 | −1/−2 | −4/−16 | −2/−8 |
| 215074_at | Myosin 1B; MYO1B; 3774 motor activity; 3779 actin binding; 5516 calmodulin binding; 5524 ATP binding. | 2q12-q34 | 5/32 | 1/2 | 4/16 |
| 220559_at | Engrailed homolog 1; 1501 skeletal development; 6355 regulation of transcription;, DNA-dependent; 9653 morphogenesis; 5634 nucleus; 3700 transcription factor activity; Protein domains-a.4.1 Homeodomain-All alpha proteins; DNA/RNA-binding 3-helical bundle; Homeodomain-like. | 2q13-q21 | 6/64 | 2/4 | 4/16 |
| 215508_at | BUB budding uninhibited benzimidazole 1 homolog (yeast); BUB1 HGNC; 910 cytokinesis;6468 protein amino acid phosphorylation; 7049 cell cycle; 7067 mitosis; 7094 mitotic spindle checkpoint; 8283 cell proliferation; 776 kinetochore; 5634 nucleus; 5816 spindle pole body; 4674 protein serine/threonine kinase activity; 5524 ATP binding; 16740 transferase activity; Pathway: Cell_cycle_KEGG GeNMAPP/KEGG. | 2q14 | 6/64 | 0/1 | 0/1 |
| 210346_s_at | CDC-like kinase 1/CLK1 HGNC; regulation of cell cycle, cell proliferation; 4674 nucleus protein serine/threonine kinase activity; 4715 non-membrane spanning protein tyrosine kinase activity; 5524 ATP binding; 16740 transferase activity; 4672 pkinase; protein kinase activity; 3.5e-82. Pathway: phosphotidylinositol signaling system GENMAPP/KEGG. | 2q33 | 0/1 | −2/−4 | 2/4 |
| 202019_s_at | Lantibiotic synthetase component C-like/ LANCL1 HGNC | 2q33-q35 | −2/−4 | −3/−8 | −2/−4 |
| 210495_x_at | Fibronectin 1: FN1 HGNC; 6953 acute-phase response; 7155 cell adhesion; 9611 response to wounding; 16477 cell migration; 5576 extracellular; 5201 extracellular matrix structural constituent; 5518 collagen binding; 8201 heparin binding. Pathway: Inflammatory_Response_Pathway GENMAPP/KEGG. | 2q34 | −2/−4 | −3/−8 | −2/−4 |
| 215151_at | DOCK 10 HGNC; dedicator of cytokinesis 10; overlaps with ABO 14594 NCBI. Homo sapiens mRNA for KIAA0694 protein, partial cds. | 2q36.3 | 7/128 | 4/16 | 7/128 |
| 216368_s_at | Collagen, type 1V, alpha 3 (Goodpasture antigen/COL4A3 HGNC; induction of apoptosis, caspase activation, cell adhesion, cell surface receptor linked signal transduction, perception of sound, circulation, cell proliferation, negative regulation of cell proliferation and angiogenesis, integrin binding, metalloendopeptidase inhibitor activity. | 2q36-q37 | 5/32 | 4/16 | 4/16 |
| 211452_x_at | Leucine rich repeat (FL11) interacting protein 1:LRRF1P1HGNC; 6357 regulation of transcription from Pol 11 promoter; 16481 negative regulation of transcription; 5634 nucleus; 5856 cytoskeleton; 3702 RNA polymerase 11 transcription factor activity; 3725 double-stranded RNA binding; 16564 transcriptional repressor activity. | 2q37.3 | 5/32 | 4/16 | 4/16 |
| 215212_at | CDNA FLJ12091 fis, clone HEMBBID02582; Hs.549985 NCBI | Align: chr 3:180376149-180378570(+)UCSC | 9/512 | 10/1024 | 5/32 |
| 207272_at | Zinc finger protein 80 (pT17); ZNF80 HGNC; 6350 Transcription; 6355 regulation of transcription, DNA-dependent; 5634 nucleus; 4700 transcription factor activity; 8270 Zinc ion binding. | 3p12-qter | 6/64 | 7/128 | 2/4 |
| 213024_at | TATA element modulatory factor 1; RMF1 HGNC; 6355 regulation of transcription, DNA-dependent; 6366 transcription from Pol 11 promoter; 3677 DNA binding; 3702 RNA polymerase II transcription factor activity; 3712 transcription cofactor activity. | 3p21-p12 | −1/−2 | −1/−2 | −1/−2 |
| 207794_at | Chemokine (C-C) receptor 2; CCR2 HGNC; 6935 chemotaxis; 6954 inflammatory response; 6968 cellular defense response; 7186 G-protein coupled receptor protein signaling pathway; 7194 negative regulation of adenylate cyclase activity; 7204 cytosolic calcium ion concentration elevation; 7259 JAK-STAT cascade; 19735 antimicrobial humoral response (sensu Vertebrata); 5625 soluble fraction; 5887 integral to plasma membrane; 1584 rhodopsin-like receptor activity; 16493 C-C chemokine receptor activity. Pathway PCRs_Class_A_Rhodopsin-like & Peptide_GPCRs GENMAPP/KEGG; | 3p21 | −1/−2 | −1/−2 | −1/−2 |
| 202009_at | PTK9L protein tyrosine kinase 9-like (A6-related protein); PRTK9L HGNC; 5622 intracellular 3779 actin binging; 5524 ATP binding; 16301 kinase activity. | 3p21.1 | −3/−8 | −2/−4 | −1/−2 |
| 201096_s_at | ADP-ribosylation factor 4/ARF4 HGNC | 3p21.2-p21.1 | −3/−8 | −2/−4 | −1/−2 |
| 39966_at | Chondroitin sulfate proteoglycan 5 (neuroglycan C)/CSPG5 HGNC; intracellular transport, neurogenesis; intracellular transport; Golgi vesicle membrane; membrane fraction; integral to plasma membrane; protein binding; growth factor activity. | 3p21.3 | 5/32 | 4/16 | 4/16 |
| 242591_at | Heat shock regulated 1; XLHSRF-1HGNC; 7018 microtubule-based movement; 9612 response to mechanical stimulus; 30286 dynein complex; 3777 microtubule motor activity. | 3p21.31 | −1/−2 | −1/−2 | −3/−8 |
| 242243_at | TATA element modulatory factor 1; TMF1 HGNC; 6355 regulation of transcription, DNA-dependent; 6366 transcription from Pol 11 promoter; 3677 DNA binding; 3702 RNA polymerase II transcription factor activity; 3712 transcription cofactor activity. | 3p21-p12 | −1/−2 | −1/−2 | −1/−2 |
| 212308_at | Cytoplasmic linker associated protein 2; CLASP2 HGNC; 7076 mitotic chromosome condensation; 5488 binding. Protein Domain: Pfam-IPRO10705 mast, C-terminal EMBL-EBI. | 3p23 | 6/64 | 3/8 | 1/2 |
| 201972_at | ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A ATPase; ATP6V1A HGNC; 6810 transport; 15986 ATP synthesis coupled proton transport; 15992 proton transport; 5887 integral to plasma membrane; 16469 proton-transporting two-sector ATPase complex; 5524 ATP binding; 16787 hydrolase activity; 16820 hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances; 46933 hydrogen-transporting ATP synthase activity, rotational mechanism; 46961; hydrogen-transporting ATPase, rotational mechanism. Pathways: oxidative phosphorylation; ATP synthesis; Photosynthesis; Flagella assembly; Type 111 secretion—GENMAPP/KEGG. | 3q13.31 | −2/−4 | −3/−8 | −3/−8 |
| 216752_at | Phosphoinositol-3-kinase, regulatory subunit 4, p150; PIK3R4 HGNC; 6468 protein amino acid phosphorylation; 7076 mitotic chromosome condensation; 7126 meiosis; 4674 protein serine/threonine kinase activity; 5488 binding; 5524 ATP binding; 16740 transferase activity. E value: 0.0 | 3q21.3 | 4/16 | 7/128 | 5/32 |
| 210703_at HG U133 | Human; Position: 145481606-145485595 (+)UCSC. PRO2259 mRNA, complete cds | Align 3q24 | 4/16 | 0/1 | 3/8 |
| 207396_s_at | ALG3 HGNC Asparagine-linked glycosylation 3 homolog (yeast, α-1,3-mannosyltransferase); lethal (2) neighbor of tid; 6486 protein amino acid glycosylation; 5783 endoplasmic reticulum, 30176 integral to endoplasmic reticulum membrane; 30 mannosyltransferase activity; 16757 glycosyl transferase activity. | 3q27.3 | 1/2 | 0/1 | 1/2 |
| 207009_at | Paired-like homeobox 2b; PHOX2B HGNC; 6355 regulation of transcription, DNA dependent; 7275 development; 7399 neurogenesis; 5634 nucleus; 3700 transcription factor activity; 3712 transcription cofactor activity. | 4p12 | 6/64 | 4/16 | 5/32 |
| 222336_at | Hypothetical protein LOC 201895. Transcription Assignments-cdna Genescan: Chromosome: NCBI 35:4:39380614: 39429366:-1. | 4p14 | 5/32 | 5/32 | 5/32 |
| 213980_s_at | C-terminal binding protein 1; CTBP1 HGNC; 6468 protein amino acid phosphorylation; 6564 L-serine biosynthesis; 8285 negative regulation of cell proliferation; 19079 viral genome replication; 5634 nucleus; 5794 Golgi apparatus; 8022 protein C-terminus binding; 16491 oxidoreductase activity; 16616 ocidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as receptor; | 4p16 | 6/64 | 5/32 | 2/4 |
| 211062_s_at | Carboxypeptidase Z; CPZ HGNC/ transmembrane receptor activity; 3.4.17.10; carboxypeptidase E activity2.26e-134. | 4p16.1 | 6/64 | 5/32 | 2/4 |
| 206639_x_at | Histatin 1; HTN1 HGNC; 1503 ossification; 6805 xenobiotic metabolism; 42742 defense response to bacteria; 50832 defense response to fungi; 5576 Extracellular region. | 4q13 | 3/8 | 3/8 | 3/8 |
| 219667_s_at | B-cell scaffold protein with ankyrin repeats 1; BANK1 HGNC/ | 4q24 | 3/8 | 6/64 | 6/64 |
| 216720_at | Cytochrome P450, family2, subfamily U, polypeptide 1 CYP2U1 HGNC; 16712 oxidoreductase activity, acting on paired donors, with incorporation of or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen. | 4q25 | 5/32 | 6/64 | 4/16 |
| 207928_s_at | Glycine receptor, alpha 3; GLRA3 HGNC; GABA-A receptor activity; ion channel activity; extracellular ligand-gated ion channel activity, glycine binding (receptor, alpha 3 subunit), glutamate–gated chloride channel activity; neurotransmitter receptor activity; synaptic transmission. | 4q33-q34 | 6/64 | 10/1024 | 8/256 |
| 213963_s_at | Sin3-associated polypeptide, 30kDa; histone deacetylase complex; transcription corepressor activity. | 4q34.1 | 4/16 | 3/8 | 1/2 |
| 215378_at | Ankyrin repeat and KH domain containing 1; ANKHD1 HGNC; nucleic acid binding; cell cycle inhibitor p16ink4A; immunoglobulin heavy chain variable domain, VH; transcription factor NusA, receptor, different EGF domains; Class II MHC alpha chain, C-terminal domain; Silencer of death domains, Sodd (Bag 4); Staphylokinase. | 5q31.3 | 2/4 | 2/4 | 2/4 |
| 207182_at | Gamma-aminobutyric acid (GABA) A receptor, alpha 6 (GABRA6 HGNC) | 5q34 | 3/8 | 8/256 | 3/8 |
| 203812_at | Slit homolog 3 (Drosophila; SLIT3 HGNC); 7399 neurogenesis, 5615 extracellular space; 5509 calcium ion binding, 5515 protein binding. | 5q35 | 0/1 | −1/−2 | −1/−2 |
| 207471_at | Representative Public ID AF 118086 NCBI; Grade A annotation; Transcript Assignment(s): AF118086 NCBI Homo sapiens PRO1992 mRNA, cds. 11/11 Chr6: 88330288-88332009(+) UCSC; unknown protein. | Align chr 6 | 9/512 | 7/128 | 5/32 |
| 202401_s_at | Serum response factor (c-fos serum response element-binding transcription factor)/SRF HGNC | 6p21.1 | −1/−2 | −1/−2 | 0/1 |
| 209823-x_at | Overlaps with Major histocompatibility complex, class II, DQ β1 (HLA-DQβ1HGNC); 6955 immune response; 19884 antigen presentation, exogenous antigen via MHC class II; 16021 integral to membrane. 45012 MHC class II receptor activity. | 6p21.3 | −1/−2 | −1/−2 | 0/1 |
| 203290_at | HLA-DQA1 HGNC; major histocompatibility complex, class II, DQ alpha 1 6955 immune response; 19884 antigen presentation, exogenous antigen via MHC class II; 5887 integral to plasma membrane; 16021 integral to membrane; 45012 MHC class II receptor activity. | 6p21.3 | −1/−2 | −1/−2 | 0/1 |
| 210208_x_at | HLA-B associated transcript 3; BAT3 HGNC. | 6p21.3 | −1/−2 | −1/−2 | 0/1 |
| 214509_at | Histone 1, H31. Transcripts alignment(s)-NM_003533NCBI-Homo sapiens histone 1, H3i (HIST1 H31), mRNA 11/11 | 6p22-p21.3 | 4/16 | 0/1 | 0/1 |
| 207156_at | Alignments: chr6: 27947602-27948078(−) UCSC. H2ag histone 1/HIST1H2AG HGNC; DNA binding; 1.2e-53; H2A histone family, | 6p22.1 | 3/8 | 5/32 | 7/128 |
| 200890-s-at | Signal signal receptor, alpha (translocon-associated protein alpha)/SSR1 HGNC) | 6p24.3 | 0/1 | 0/1 | 0/1 |
| 214790_at | SUMO1/sentrin specific protease 6 SENP6 HGNC; 6508 proteolysis and peptidolysis; 6512 ubiquitin cycle; 8234 cysteine-type peptidase activity. | 6q13-q14.3 | 4/16 | 5/32 | 3/8 |
| 207054_at | Interphotoreceptor Matrix Proteoglycan 1; IMPG1 HGNC; 7601 visual perception; 5578 Extracellular matrix (sensu metazoa); 4872 Receptor activity; 5201 Extracellular matrix structural constituent. | 6q14.2-q15 | 6/64 | 4/16 | 8/256 |
| 211387_x_at | RNA guanylyltransferase and 5′-phosphatase; RNGTT HGNC; mRNA capping, protein amino acid dephosphorylation, nucleic acid binding, transferase activity, hydrolase activity. Tyrosine specific protein phosphatase and dual specificity protein phosphatase. | 6q16 | 3/8 | 3/8 | −1/−2 |
| 205029_s_at | Fatty acid binding protein 7, brain; FABP7 HGNC; 6631 FA metabolism; 6810 transport; 7399 neurogenesis; 8285 negative regulation of cell proliferation; 5215 transporter activity; 5478 Intracellular transporter activity; 8289 lipid binding. | 6q22-q23 | 7/128 | 5/32 | 8/256 |
| 217829_s_at | Peptidylprolyl isomerase A (cyclophilin A); protein folding; antimicrobial humoral response (sensu Vertebrata); regulation of viral genome replication; chaperone activity; peptidyl-prolyl cis-trans isomerase activity; protein transporter activity. | 7p13-p11.2 | 4/16 | 0/1 | 3/8 |
| 201293_x_at | Peptidylprolyl isomerase A (cyclophilin A); PPIA HGNC; 6457 protein folding; 19735 antimicrobial humoral response (sensu Vertebrata); 45069 regulation of viral genome replication; 3754 chaperone activity; 3755 peptidyl-prolyl cis-trans isomerase activity; 8565 protein transporter activity; 16853 isomerase activity. Alignment with chromosomes 1–3, 5–21, and X. | 7p13-p11.2 | 4/16 | 0/1 | 3/8 |
| 208641_s_at | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1); RAC1 HGNC; 6928 cell motility; 6954 inflammatory response; 7155 cell adhesion; 7264 small GTPase mediated signal transduction; 9653 morphogenesis; 3924 GTPase activity; 5525 GTP binding; 3925 ras; small monomeric GTPase activity;3e-92 extended.;2.5e-84 extended. | 7p22 | 7/128 | 4/16 | 2/4 |
| 206202_at | Mesenchyme homeobox 2 (growth arrest specific homeobox); MEOX2 HGNC; 6355 regulation of transcription, DNA-dependent; 7275 development; 8015 circulation; 5634 nucleus; 3700 transcription factor activity. | 7p22.1-p21.3 | 7/128 | 4/16 | 2/4 |
| 207166_at | Gamma transducing activity polypeptide 1 guanine nucleotide binding protein (G protein); GNGT1 HGNC/G-gamma; heterotrimeric G-protein GTPase activity; 2.8e-26; gamma transducing activity polypeptide 1; photoreceptor transduction gamma subunit (fragment). | 7q21.3 | 5/32 | 7/128 | 2/4 |
| 206552_s_at | Tachykinin, precursor 1(substance K, substance P, neurokinin 1 & 2, neuromedin L, neurokinin α, neuropeptide K, neuropeptide γ/TAC1 HGNC; 7217tachykinin signaling pathway; 7218 neuropeptide signaling pathway; 7267 cell-cell signaling; 7268 synaptic transmission; 7320 insemination; 9582 detection of abiotic stimulus; 5102 receptor binding; 5184 neuropeptide hormone activity. | 7q21-q22 | 5/32 | 0/1 | 4/16 |
| 215516_at | Laminin, beta 4 LAMB4 HGNC. 5578 Extracellular matrix (sensu Metazoa); 5198 Structural molecule activity. Protein Domain-Pfam IPP008211-Laminin, N-terminal EMBL-EBI; Pfam IPR002049-Laminin-type EGF-like domain EMBL-EBI. | 7q22-q31.2 | 5/32 | 6/64 | 2/4 |
| 209466_x_at | Pleiotrophin (heparin binding growth factor 8, neurite growth promoting factor 1); neurotrophic factor/PTN HGNC; osteoblastic specific factor-1, OSF-1. | 7q33-q34 | 1/2 | 0/1 | 0/1 |
| 207166_at | Potassium voltage-gated channel, subfamily H(eag-related, member 2/KCNH2 HGNC; | 7q35-q36 | 5/32 | 7/128 | 2/4 |
| 205262_at | 6812 cation transport; 160 two component signal transduction system (phosphorylase) 6813 potassium ion transport; 6936 muscle contraction, 7605 perception of sound, 8016 regulation of heart rate; 6355 PAC; regulation of transcription, DNA dependent; 9l2e-09; 5624 membrane fraction; 8076 voltage-gated potassium channel complex; 16021 integral membrane; 155 two-component sensor molecule activity;5251 delayed rectifier potassium channel activity; 5216 ion_trans; ion channel activity; 6.1e-31. | 7q35-q36 | 5/32 | 7/128 | 2/4 |
| 209441_at | Rho-related BTB domain containing 2/RHO BTB2HGNC; ras, small monomeric GTPase activity; 1.3e-16; 5515 protein binding; 5525 GTP binding | 8p21.2 | 5/32 | 3/8 | 3/8 |
| 207987_s_at | Gonadotropin-releasing hormone 1(Leutenizing-releasing hormone; GNR1 HGNC; signal transduction, cell-cell signaling, development, negative regulation of cell proliferation; leutenizing; GnRH. | 8p21-p11.2 | 5/32 | 2/4 | 1/2 |
| 219312_s_at | Zinc finger and BTB domain containing 10 ZBTB10 HGNC; 6350 transcription; 6355 regulation of transcription, DNA-dependent; 5634 nucleus; 3677 DNA binding; 5515 protein binding; 8270 Zinc ion binding. E value: 0.0 | 8q13-q21.1 | 3/8 | 2/4 | 2/4 |
| 204865_at | Carbonic anhydrase III, muscle specific; CA3 HGNC; 6730 one carbon compound metabolism; 5737 cytoplasm; 4089 carbonic anhydrase activity; 8270 Zinc ion binding; 16829 Lyase activity; Pathway: Nitrogen metabolism GENMAPP/KEGG. | 8q13-q22 | 6/64 | 3/8 | 4/16 |
| 209066_x_at | Ubiquinol-cytochrome c reductase binding protein; UQCRB HGNC; 8121 UCR_14kD; ubiquinol-cytochrome-c reductase activity;4.7e-56. Pathway: Oxidative phosphorylation; | 8q22 | 6/64 | 3/8 | 4/16 |
| 205528_s_at | Electron_Transport_Chain GENMAPP/KEG. Runt-related transcription factor 1; translocated to, 1(cyclin D-related); RUNX 1TI HGNC; 6091 generation of precursor metabolites and energy; 6350 transcription; 6355 regulation of transcription, DNA-dependent; 5634 nucleus; 3700 transcription factor activity; Protein Domain: Pfam: IPR 000040 EMB-EBI Acute myeloid leukemia 1 protein (AML1)/RUNT; IPR 002893 EMB-EBI Zn-finger, MYND type. | 8q22 | 5/32 | 1/2 | 3/8 |
| 200640_at | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide/YWHAZ HGNC; 19904 protein domain specific binding; 14-3-3 protein/cytosolic phospholipase A2 (Fragmnet); alignment(s) chr 10, 15, 2, 6, 8,9 and x. | 8q23.1 | −4/−16 | −5/−32 | −4/−16 |
| 244089_at | v-myc myelocytomatosis viral oncogen homolog(avian); MYC HGNC; 7050 cell cycle arrest; 6879 iron ion homeostasis, 8283 cell proliferation; 5634 nucleus; 6357 regulation of transcription from Pol II promoter; 3700 transcription factor activity; Pathway: Apoptosis GENMAPP/KEGG; Wnt_signaling GENMAPP/KEGG. | 8q24.12-q24.13 | 1/2 | 0/1 | 1/2 |
| 212654_at | Tropomyosin 2 (beta)/TPM2 HGNC; muscle development; cytoskeleton; muscle thin filament tropomyosin. | 9p13.2-p13.1 | 6/64 | 2/4 | 3/8 |
| 207932_at | Interferon, alpha 8 IFNA 8HGNC; 6952 Defense response; 5576 extracellular region; 5126 hematopoietin/interferon-class (D200-domain) cytokine receptor binding. E value 1.0E-102. | 9p22 | 5/32 | 4/16 | 3/8 |
| 209819_at | Hyaluronan binding protein 4 HABP4 HGNC Annotation transcript cluster (# of matching probes) NM_014282(11), u77327 (11); Transcript Annotations- u77327 NCBI; NM 014282- Human Ki-1/57 intracellular antigen mRNA, partial cds. 11/11 A. Homo sapiens hyaluronan binding protein 4 (HABP4) 11/11 | 9q22.3-q31 | 4/16 | 4/16 | 3/8 |
| 202491_s_at | Kinase complex-associated protein inhibitor kappa light polypeptide gene enhancer in B-cells; IKBKAP HGNC; 6461 protein complex assembly; 6468 protein amino acid phosphorylstion; 6955 immune response; 4871 signal transducer activity; 8607 phosphorylase kinase regulator activity; 16301 kinase activity. | 9q31 | −2/−4 | 0/1 | −4/−16 |
| 211402_s_at | Nuclear receptor subfamily 6, group A, member 1, NR6A1 HGNC; nuclear hormone receptor; transcription factor activity; regulation of transcription, spermatogenesis, cell proliferation, steroid hormone receptor activity, Err2; Orphan nuclear receptor NR5a2 (LRH-1). | 9q33-q34.1 | 1/2 | 0/1 | 1/2 |
| 218813_s_at | SHE-domain GRB2-like endophilin B2/SH3GLB2 HGNC. | 9q34 | 4/16 | 2/4 | 4/16 |
| 222241_at | chromosome 9 open reading frame 28; C9orf28 HGNC;(Full length) Hs.438972 NCBI; LocusLink 89853 Entrez gene. | 9q34 | 4/16 | 2/4 | 4/16 |
| 217709_at | N-myristoyltransferase 2 NMT2 HGNC; EC 2.3.1.97; 6499 N-terminal protein myrstolation; 9249 protein lipolation; 4379 glycylpeptide N-tetradecanoyltransferase activity; 8415 acyltransferase activity; 16740 transferase activity. | 10p13 | 8/256 | 2/4 | 5/32 |
| 219433_at | 5-OHtryptamine (serotonin) receptor 7 (adenylate-cyclase- coupled): G-protein signaling, coupled to cyclic nucleotide second messenger; synaptic transmission; circadian rhythm; circulation; Cellular component: integral to plasma membrane; rhodopsin-like receptor activity; melanocortin receptor activity; serotonin receptor activity. | 10q21-q24 | 6/64 | 2/4 | 1/2 |
| 216098_s_at | 5-hydroxytryptamine (serotonin) receptor 7 (adenylate cyclase-coupled) serotonin-7 receptor pseudogene /HTR7, LOC 93164 HGNC; 7187 G-protein signaling, coupled to cyclic nucleotide second messenger; 7268 synaptic transmission; 7623 circadian rhythm; 8015 circulation; 5887 integral to plasma membrane; 1584 rhodopsin-like receptor activity; 4993 serotonin receptor activity; 4977 melanocortin receptor activity. Pathway: GPCRDB_Class_A_Rhodopsin-like; momoamine_GPCRs GENMAPP/KEGG | 10q21-q24 | 8/256 | 6/64 | 4/16 |
| 210110_x_at | Heterogenous nuclear ribonucleoprotein H3 (2H9) HNRPH3 HGNC; 6397 mRNA processing; 5634 nucleus; 30529 ribonucleoprotein complex; 3723 RNA binding; 3676 nucleic acid binding; 8e-12 extended. | 10q22 | 8/256 | 6/64 | 4/16 |
| 202361_at | SEC24 related gene family, member C (S. cerevisiae); SEC24C HGNC; 6886 intracellular protein transport; 6888 EDR to Golgi transport; 5783 endoplasmic reticulum; 5794 Golgi apparatus; 30127 COP11 vesicle coat; 3779 actin binding; protein binding; 8565 protein transporter activity. | 10q22.3 | 8/256 | 6/64 | 4/16 |
| 207661_s_at | SH3 multiple domains 1; SH3MD1 HGNC; | 10q25.1 | 3/8 | 5/32 | 2/4 |
| 205493_s_at | Dihydropyrimidinase-like 4; DPYSL4 HGNC; 7399 neurogenesis; methylenetetrahydrofolate dehydrogenase/ cyclohydrolase; 16787 hydrolase activity. | 10q26 | 8/256 | 6/64 | 8/256 |
| 201458_s_at | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast)/BUB3 HGNC; 7067 mitosis; 7094 mitotic spindle checkpoint; 8283 cell proliferation; 776 kinetochore; 5634 nucleus. Pathway: Cell_cycle GENMAPP/KEGG. | 10q26 | −1/−2 | −3/−8 | −3/−8 |
| 208229_at | Fibroblast growth factor receptor (FGF) 2 (bacterial espressed kinase, keratinocyte growth factor receptor; craniofacial dysostosis 1, Crouzon syndrome, Pfeiffer syndrome, Jackson-Weiss syndrome)/FGFR2 HGNC; 6468 protein amino acid phosphorylation; 16021 integral to membrane; 4713 protein-tyrosine kinase activity; 4872 receptor activity; 5007 FGF receptor activity; 5524 ATP binding; 16740 transferase activity. | 10q26 | 3/8 | 0/1 | 0/1 |
| 221950_at | Empty spiracles homolog 2(Drosophila) EMX2 HGNC; Homeobox protein EMX1; transcription factor activity; regulation of transcription, DNA- dependent. | 10q26.1 | 5/32 | 8/256 | 4/16 |
| 221127_s_at | Regulated in glioma; RIG HGNC; aligns with 11p15.3 | 11p15.1 | 5/32 | 4/16 | 2/4 |
| 208607_s_at | Serum amyloid A2; SAA2 HGNC 6953 acute phase response; 6954 inflammatory response; 5576 extracellular; 5319 lipid transporter activity; 3794 SAA_proteins; acute phase response protein activity; 1.3e-79. Serum amyloid A-2 protein precursor [contains: Amyloid protein A (Amyloid fibril protein AA)]. | 11p15.1-p14 | 5/32 | 4/16 | 2/4 |
| 200023_s_at | 47kDa eukaryotic translation initiation factor 3, subunit 5 epsilon; EIF3S5 HGNC; 6412 protein biosynthesis; 6446 regulation of translational initiation; 3743 translational initiation factor activity. Pathway: Translational_Factors GENMAPP/KEGG. | 11p15.4 | −3/−8 | −6/−64 | −6/−64 |
| 210538_s_at | Baculoviral IAP repeat-containing 3, BIRC3 HGNC; 6916 anti apoptosis; 7166 cell surface linked signal transduction; 16567 protein ubiquination; 42981 regulatio of apoptosis; 151 ubiquitin ligase complex; 4842 ubiquitin-protein ligase activity; 5515 protein binding; 8270 zinc ion binding; Pathway: Apoptosis GENMAPP/KEGG | 11q22 | 6/64 | 6/64 | 4/16 |
| 215355_at | POU domain, class 2, transcription factor 3/POU2F3 HGNC; regulation of transcription, DNA-dependent | 11q23.3 | 7/128 | 5/32 | 4/16 |
| 220611_at | Male sterility containing 1 MLSTD1 HGNC | 12p11.23 | 2/4 | 4/16 | 4/16 |
| 220615_s_at | Male sterity domain; MLSTD HGNC/ PGDH Human- EC: 1.1.1.141:15-Hydroxyprostaglandin Dehydrogenase [NAD (+)]. | 12p11.23 | 0/1 | −3/−8 | −1/−2 |
| 213764_s_at | Microfibrillar associated protein 5; MFAP5 HGNC; 1527 microibril; 5578 extracellular matrix (sensu Metazoa); 5615 extracellular space; 5201 extracellular matrix structural constituent. | 12p13.1- p12.3 | 9/512 | 6/64 | 5/32 |
| 201604_s_at | Protein phosphotase 1, regulatory (inhibitor) subunit 12A/ PPP1R12A HGNC; myosine phosphotase, target subunit 1 [Homo sapiens] | 12q15-q21 | 2/4 | 1/2 | 1/2 |
| 208752__x_at | Nucleosome assembly protein-1-like 1/NAP1L1 HGNC; 6260 DNA replication; 6334 nucleosome assembly; 8284 positive regulation of cell proliferation; 5634 nucleus; 5678 chromatin assembly complex; 3677 DNA binding. | 12q21.1 | 1/2 | 1/2 | 0/1 |
| 216466_at | Neuron navigator 3; nucleotide binding; pore membrane and/or filament interacting like protein 1; steerin 3 [homo sapiens]; Calponin-like actin-binding; rhodopsin-like GPCR superfamily ATPase | 12q21.2 | 7/128 | 2/4 | 4/16 |
| 204840_s_at | Early Endosome Antigen 1, 162kD; EEA1 HGNC; 6906 vesicle fusion; 16189 synaptic vesicle to Endosomal fusion; 45022 Early Endosome to late endosome transport; 5624 membrane fraction; 5634 nucleus 5769 Early Endosome; 5829 cytosol; 19897 Extrinsic to plasma membrane; 3676 nucleic acid binding; 5516 Calmodulin binding; 5545 phosphotidylinositol binding; 8270 Zinc ion binding; 30742 GTP-dependent protein binding; 42803 protein homodimerization activity | 12q22 | 2/4 | 5/32 | 2/4 |
| 201523_x_at | Ubiquitin-conjugating enzyme E2N (UBC13 homolog, yeast) HGNC); 6282 regulation of DNA repair; 6464 protein modification; 6508 proteolysis and peptidolysis; 4840 ubiquitin conjugating enzyme activity; 4842 ubiquitin-protein ligase activity. Pathway: Ubiquitin mediated proteolysis GENMAPP/KEGG. | 12q22 | −3/−8 | 0/1 | −2/−4 |
| 210515_at | Transcription factor 1, hepatic; LF-B1, hepatic nuclear factor (HNF1), albumin proximal factor transcription factor 1; RNA polymerase 11 transcription factor activity. | 12q24.2 | 4/16 | 0/1 | 1/2 |
| 207956_x_at | Androgen-induced proliferation inhibitor; APRIN HGNC; androgen-induced prostate proliferative shutoff associated protein; 6355 regulation of transcription, DNA-dependent 8283 cell proliferation; 8285 negative regulation of cell proliferation; 5634 nucleus; 3677 DNA binding; 5524 ATP binding. | 13q12.13 | −3/−8 | −1/−2 | −3/−8 |
| 204190_at | Chromosome 13 open reading frame 22; C13orf22 HGNC/ 4197 cysteine-type endopeptidase activity; 4221 ubiquitin thioesterase activity; | 13q12-q14 | 2/4 | 2/4 | 2/4 |
| 219471_at | Chromosome 13 open reading frame 18; C13orf18 HGNC; (Full length-UniGene ID Build 170 ()Hs.413071 NCBI. | 13q14.11 | 2/4 | 2/4 | 2/4 |
| 211119_at | Estrogen receptor 2 (ER beta); regulation of transcription, DNA-dependent; signal transduction; cell-cell signaling; negative regulation of cell growth; steroid-binding; infertility. Ribosomal protein S29; RPS29 HGNC; 3735 Ribosomal_S14; structural constituent of ribosome; 2.7e-13 extended. | 14q | 8/256 | 5/32 | 8/256 |
| u133A 201094_at | Zinc finger and BTB domain containing 1; ZBTB1 HGNC/DNA binding; protein binding, zinc ion binding. | 14q23.3 | 5/32 | 2/4 | 5/32 |
| 205092_x_at | Zinc finger and BTB domain containing 1; DNA- binding; protein binding; zinc ion binding: regulation of transcription, DNA-dependent. | 14q23.3 | 5/32 | 2/4 | 5/32 |
| 220726_at | Chromosome 14 open reading frame 157/C14orf157 HGNC. | 14q32.33 | 11/2048 | 11/2048 | 8/256 |
| 220726_at | Chromosome 14 open reading frame 157/C14orf157 HGNC. Representative Public ID- NM_024100 NCBI. Target transcription: gb: NM_025100.1/DEF= Homo sapiens hypothetical protein FLJ 12294/PROD= hypothetical protein etc. | 14q32.33 | 11/2048 | 11/2048 | 8/256 |
| 216892_at | Immunoglobulin heavy locus/IGH@ HGNC; IGHV3 protein (Fragment); CDNA FLJ25997 fis, clone DMC06923, highly similar to Ig gamma-1chain EMBL-EBI C region. | 14q32.33 | 2/4 | 1/2 | 2/4 |
| 214328_s_at | Heat shock 90kDa protein 1, alpha; 6467 protein folding; 6839 mitochondrial transport; 7165 signal transduction; 42026 protein refolding; 45429 positive regulation of nitric oxide biosynthesis; 5829 cytosol; 166 nucleotide binding; 3754 chaperone activity; 3773 heat shock protein activity; 5515 protein binding; 5524 ATP binding; 30235 nitric oxide synthase regulator activity; 30911 TPR domain binding; 42803 protein homodimerization activity. | 14q32.33 | 2/4 | 1/2 | 2/4 |
| 219196_at | Secretogranin III SCG3 HGNC; Transcript alignment(s)- NM_013243 NCBI-Homo sapiens secretogranin III (SCGS), mRNA. | 15q21 | 9/512 | 8/256 | 9/512 |
| 209830_s_at | Solute carrier family 9(sodium/hydrogen) exchanger, isoform 3 regulator 2/SLC9A3R2 HGNC; 6461 protein complex assembly; 7242 intracellular signaling cascade; 5634 nucleus; 5886 plasma membrane; 5515 protein binding; blast- embl CAA90511.1/tyrosine kinase activating protein 1 (TKA-1) Homo Sapiens. | 16p13.3 | 7/128 | 6/64 | 4/16 |
| 207756_at | Transcript Assignment(s) AKO 24372 NCBI- Homo sapiens cDNA FLJ14310fis, clone PLACE 3000271. Alignment(s) –chr16: 45516108–45519963(+) UCSC. Protein Similarities: blast P39189-Alu subfamily SB sequence contamination warning entry; blast AAG23169- HC6 (Homo sapiens) | Align Chr 16 | 7/128 | 3/8 | 7/128 |
| 216623_x_a | Trinucleotide repeat containing 9; TNRC9 HGNC; 785 chromatin; 5634 nucleus; 3677 DNA binding; blast- trinucleotide repeat containing 9 (Mus masculine). | 16q12.1 | 8/256 | 6/64 | 9/512 |
| 211456_x_at | Metallothionein 1H; 46872 metal ion binding. | 16q13 | 7/128 | 6/64 | 4/16 |
| 220575_at | Hypothetical protein FLJ11800 HGNC; alignment Ch 17 p11.2 / q23.1 | Alignment Ch 17 p11.2 / q23.1 | 4/16 | 3/8 | 4/16 |
| 207704_s_at | Growth arrest-specific 7/GAS HGNC; 7050 cell cycle arrest; 7275 development; 7399 neurogenesis; 8151 cell growth and/or maintenance; 3700 transcription factor activity: Database ID d1srda_SCOP:b.1.8.1:| Cu, Zn superoxide dismutase, SOD. | 17p13.1 | 1/2 | 1/2 | 2/4 |
| 210816_s_at | Cytochrome b-561/CYB561HGNC; energy pathways, 6118 electron transport; integral to plasma membrane; 4128 cytochrome-b5 reductase activity;/cytochrome b561/ferric reductase transmembrane. | 17q11-qter | 0/1 | −1/−2 | −1/−2 |
| 200614_at | Heavy polypeptide (Hc) clathrin; CLTC HGNC; 6886 intracellular protein transport;5902 coated pit; 30125 clathrin vesicle coat; 5198 structural molecule activity. | 17q11-qter | 0/1 | −1/−2 | −1/−2 |
| 200029_at | Ribosomal protein L19(60S); RPL19 HGNC; 6412 protein synthesis; 5622 intracellular; 5840 ribosome; 5842 cytosolic large ribosomal subunit (sensu Eudarya); 3723 RNA binding; 3735 structural constituent of ribosome; Pathway-Ribosomal_Proteins GENMAPP/KEGG. | 117q11-qter q11-qter | 0/1 | −1/−2 | −1/−2 |
| 204995_at | Cyclin-dependent kinase 5, regulatory subunit 1(p35); CDK5R1 HGNC;cyclin-dependent protein kinase 5 activator complex; protein kinase activity, | 17q12 | 3/8 | 1/2 | 2/4 |
| 211310_at | Enhancer of zeste homolog 1(Drosophila; EZH1 HGNC; 6350 transcription; 6355 regulator of transcription, DNA dependent; 9653 morphogenesis; 5634 nucleus; 3682 chromatin binding. | 17q21.1- q21.3 | 6/64 | 6/64 | 5/32 |
| 205366_s_at | Homeo box B6; HOX HGNC; 6355 regulation of transcription, DNA-dependent; 7275 development; 8595 determination of anterior/posterior axis, embryo; 5634 nucleus; 3700 transcription factor activity. | 17q21.3 | 2/4 | 2/4 | 2/4 |
| 208430_s_at | Dystrobrevin, alpha/DTNA HGNC; striated muscle contraction; signal transduction; neuromuscular synaptic transmission, cell growth and/or maintenance; 5515 protein binding; 8270 zinc ion binding. | 18q12 | 3/8 | 2/2 | 0/1 |
| 218738_s_at | Ring finger protein 138; RNF 138 HGNC; (Full length) Hs.180403 UniGene ID Build 170 (); 5634 nucleus; 3676 nucleic acid binding; 8270 zinc ion binding | 18q12.1 | 3/8 | 2/4 | 0/1 |
| 207315_at | CD226 antigen; CD226 HGNC; cell adhesion, signal transduction, antimicrobial humoral response (sensu Vertebrata); integral to plasma membrane; NK cell activating receptor NKP44; Myelin oligodendrocyte glycoprotein (MOG); immunoglobulin-like. | 18q22.3 | 2/4 | 4/16 | 2/4 |
| 203022_at | Large subunit ribonuclease H2; RNASEH2A HGNC/ DNA replication, RNA catabolism, RNA binding, endonuclease activity, hydrolase activity, ribonuclease activity. | 19p13.2 | 0/1 | −1/−2 | −1/−2 |
| 205382_s_at | D component of complement (adipsin)/DF HGNC 6508 proteolysis and peptidolysis; 6957 complement activation, alternative pathway; 3817 complement factor D activity; 4263 chymotrypsin activity; 4295 trypsin activity; 8233 peptidase activity; 16787 hydrolase activity; Protein Similarities: blastx P00746 Complement factor D precursor (EC 3.4.21.46) (EC 3.4.21.46)(C3 convertase activator) | 19p13.3 | 1/2 | 0/1 | 3/8 |
| 208037_s_at | MADCAM1 HGNC; mucosal vascular addressin cell adhesion molecule 1; 6955 immune response; 7155 cell adhesion; 7165 signal transduction; 5624 membrane fraction; 16021 integral to membrane; 5515 protein binding. | 19p13.3 | 1/2 | −1 | 3/8 |
| 213490_s_at | MAP2K2 HGNC; mitogen-activated protein kinase kinase 2; 6468 protein amino acid phosphorylation; 5576 extracellular; 4674 protein ser/thr kinase activity; 4713 protein-tyrosine kinase activity; 5524 ATP binding; 16740 transferase activity; 4672 protein kinase activity; 5.3e-76. Pathway Integrin-mediated cell adhesion GENMAPP/KEGG; MAPK_Cascade GENMAPP/ KEGG. | 19p13.3 | 1/2 | 0/1 | 3/8 |
| 203809_s_at | v-akt murine thymoma viral oncogene homolog 2; AKT2 HGNC; 6468 protein amino acid phosphorylation;4674 protein serine/threonine kinase activity; 5524 ATP binding; 16740 transferase activity; Pathway Integrin-mediated_cell_adhesion; A1P_Signaling GENMAPP/KEGG. | 19q13.1- q13.2 | −2/−4 | −1/−2 | 0/1 |
| 202014_s_at | Protein phosphatase 1, regulatory (inhibitor) subunit 15A; PPP1R15A HGNC; 6915 apoptosis; 6974 response to DNA damage stimulus; 7050 cell cycle arrest. | 19q13.2 | −2/−4 | −1/−2 | −1/−2 |
| 200623_s_at | Calmodulin 3 (phosphatase kinase alpha, delta; CALM3HGNC; 1539 ciliary/flagellar motility; 9288 flagellum (sensu Bacteria); 5509 calcium ion binding; 16301 kinase activity; Pathways: G13_Signaling_Pathway: G_Protein_Signaling; Glycogen_Metabolism; G13_13 Signaling_Pathway GENMAPP/KEGG. | 19q13.2- q13.3 | −2/−4 | −1/−2 | 0/1 |
| 43544_at | Thyroid hormone receptor associated protein 5; THRAP5 HGNC; 6357 regulation of transcription from Pol 11 promoter; 6366 transcription from Pol 11 promoter; 6367 transcription initiation from Pol 11 promoter; 30521 androgen receptor signaling pathway; 119 mediator complex; 5634 nucleus; 3712 transcirption cofactor activity; 3713 transcription coactivator activity; 4872 receptor activity; 16455 RNA polymerase 11 transcription mediator activity; 16563 transcriptional activator activity; 30374 ligand-dependent nuclear receptor transcription coactivator activity; 42809 vitamin D receptor binding; 46966 thyroid hormone receptor binding. | 19q13.3 | 1/2 | 0/1 | 3/8 |
| 201640_x_at | Cleft lip and palate associated transmembrane protein 1: CLPTM 1 HGNC; 7275 development; 5887 integral to membrane. | 19q13.3 | 1/2 | 0/1 | 3/8 |
| 202942_at | Beta polypeptide electron-transfer-flavoprotein; ETFB HGNC; 6118 electron transport; 5759 mitochondrial matrix; 9055 electron carrier activity; 5489 ETF beta; electron transport activity; 1.4e-128 extended. | 19q13.3 | 1/2 | 0/1 | 3/8 |
| 200003_s_at | Ribosomal protein L28/RPL28 HGNC; 3735 3.2e-80 extended; Pathway: Ribosomal_proteins GENEMAPP/ KEGG. | 19q13.4 | −4 | −7/−128 | −3/−8 |
| 207857_s_at | Member 2 leukocyte immunoglobulin-like receptor, subfamily A (with TM domain); LILRA2 HGNC; 6955 immune response; 16021 integral to membrane; 4872 receptor activity; Database d1m4ka2 SCOP: b.1.4:| Killer cell inhibitory receptor. | 19q13.4 | −4/−16 | −7/−128 | −3/−8 |
| 200024_at | Ribosomal protein S5 ; RPS5 HGNC ; 6412 protein biosynthesis ; 5622 intracellular ; 5843 cytosolic small ribosomal subunit (sensu Eukarya) ; 3723 RNA binding ; 3735 structural constituent of ribosome. | 19q13.4 | −4/−16 | −7/−128 | −3/−8 |
| 217075_x_at | Transcript Description:-Consensus sequence. Grade A annotation. Representative Public ID: AF105279 NCBI; Target Description: Consensus includes gb: AF105279. 1/DEF=Homo sapiens myeloid lymphoid leukemia 2(MLL2) mRNA, alternatively spliced, partial cds. | Align chro19: 40903643–40904271 (+) UCSC. | 7/128 | 4/16 | 7/128 |
| 207465_at | Transcript ID-g7662575. Reported Public ID NM_014134 NCBI; gb:NM_014134.1/DEF=Homo sapiensPRO0628 protein. | Align: chr 20:39099060–39100238 (−)UCSC. | 8/256 | 8/256 | 5/32 |
| 209061_s_at | Nuclear receptor coactivator 3; NCOA3 HGNC— regulation of transcription; DNA-dependent signal transduction; 6350 transcription; 6355 regulation of transcription, DNA-dependent; 7165 signal transduction; 5634 nucleus; 3713 transcription coactivator activity; 4402 histone acetyltransferase activity; 46966 thyroid hormone receptor binding. Similar to nuclear receptor coactivator 3 isoform a; amplified in breast cancer-1. | 20q12 | 1/2 | 3/8 | 4/16 |
| 205153_s_at | Tumor necrosis factor receptor superfamily, member 5; TNFRSF5 HGNC; 6461 protein complex assembly; 6915 apoptosis; 6954 inflammatory response; 6955 immune response; 7165 signal transduction; 7275 development; 19735 antimicrobial humoral response (sensu Vertebrata); 30168 platelet activation; 42100 B-cell activation; 43123 positive regulation of I-kappaB kinase/NF-kappaB cascade; 5887 integral to plasma membrane; 4888 transmembrane receptor activity; 4872 TNFR_c6; receptor activity; 2.8e-10. Pathway: Apoptosis; Inflammatory_Response_Pathway GENMAPP/ KEGG. | 20q12-q13.2 | 1/2 | 3/8 | 4/16 |
| 219568_x_at | SRY (sex determining region Y)-box 18; SOX18 HGNC; 6357 regulation of transcription from RNA polymerase II polymerase II promoter; 3677 DNA-binding; 3702 RNA polymerase II transcription factor activity; 5634 nucleus; 6350 transcription. | 20q13.33 | 2/4 | 1/2 | 1/2 |
| 222310 _at | Splicing factor, arginine/serine-rich 15; pre-mRNA splicing protein rA4 [Homo sapiens]; Regulation of nuclear pre-mRNA protein; SFRS HGNC; 3723 RNA binding; 5634 nucleus. | 21q22.1 | 1/2 | 5/32 | 3/8 |
| 211113_s_at | ATP-binding cassette, sub-family G (WHITE), member 1/ABCG1 HGNC; cholesterol homeostais, lipid transport, detection of hormone stimulus; response to organic substance; integral to plasma membrane; nucleotide binding; L-tryptophan transporter activity; purine nucleotide transporter activity; protein dimerization activity. | 21q22.3 | 5/32 | 1/2 | 1/2 |
| 219433_at | BCL6 co-repressor/BCOR HGNC; Bcl-6 interacting corepressor isoform 1, 2. nasopharyngeal carcinoma susceptibility protein LZ16 mRNA. | Xp11.4 | 6/64 | 2/4 | 2/4 |
| 219433_at u133A | BCL6 co-repressor; immunoglobulin-binding protein G, different constituent domains; Ankyrin; myotrophin | Xp11.4 | 6/64 | 2/4 | 1/2 |
| 207580_at | Melanoma antigen, family B, 4/MAGEB4 HGNC. | xp21.3 | 5/32 | 1/2 | 4/16 |
| 206148_at | Alpha (low affinity) interleukins 3 receptor; IL3RA HGNC; 6468 protein amino acid phosphorylation; 7275 development; 16021 integral to membrane; 4872 hematopoietin/interferon-class (D200-domain) cytokine receptor activity; 5057 receptor signaling protein activity. | xp22.3 or yp11.3 | −2/−4 | −1/−2 | −2/−4 |
| 203903_s_at | Hephaestin HEPH HGNC/ ferroxidase activity; Ceruloplasmin, multicopper oxidase, type 1; 5507 copper-binding site; 43221.1.16.3.1;ferroxidase activity; 1e-300 4322 1.16.3.1;ferroxidase activity; 9.87e-249. | Xq11-q12 | 3/8 | 1/2 | 0/1 |
| 202711_at | Ephrin-B1; EFNB 1 HGNC; 7155 cell adhesion; 7267 cell-cell signaling; 7275 development; 7399 neurogenesis; 5625 soluble fraction; 5887 integral to plasma membrane; 16020 Ephrin; membrane;9.e-88; 46875 ephrin receptor binding. | xq12 | 3/8 | 1/2 | 0/1 |
| 201994_s_at | Mortality factor 4 like 2/MORF4L2 HGNC; 1558 regulation of growth; 5634 nucleus. Similar to blastx Q9Y690 EMBL-EBI Transcription factor-like protein MORF4 (Mortality factor 4) (Cellular senescence- related protein 1) (SEN 1). | Xq22 | −2/−4 | −1/−2 | −1/−2 |
| 200896_s_at | Hepatoma-derived growth factor (high-mobility group protein 1 –like/HDGF HGNC; 7165 signal transduction; 8283 cell proliferation; 5615 extracellular space; 5737 cytoplasm; 8083 growth factor activity 8201 heparin binding. | Xq25 | −1/−2 | −1/−2 | −1/−2 |
| 215555_at | CDNA FLJ13712fis, clone PLACE E2000394 Hs.549748 NCBI | TBC* | 8/256 | 7/128 | 8/256 |
| 220222_at | PRO1905 HGNC; Hypothetical protein. | TBC* | 7/128 | 4/16 | 5/32 |
Acknowledgements
References
- Pier, S. M. The role of heavy metals in human health. Texas Rep Biol Med 1975, 33(1), 85–106. [Google Scholar]
- Whitekus, M. J.; Santini, R. P.; Rosenspire, A. J.; McCabe, M. J., Jr. Protection Against CD95-Mediated Apoptosis by Inorganic Mercury in Jurkat T Cells. The J. Immunol 1999, 162, 7162–7170. [Google Scholar]
- Ashkenazi, A.; Dixit, V. M. Death receptors: signaling and modulation. Science 1998, 281, 1305–1317. [Google Scholar]
- Ashkenazi, A.; Dixit, V. M. Death Receptors: Signaling and Modulation. Science 2002, 281(5381), 1305–1326. [Google Scholar]
- Tartaglia, L. A.; Ayres, T. M.; Wong, G. H. W.; Goeddel, D. V. A novel domain within the 55 kD TNF receptor signals cell death. Cell 1993, 74, 45–53. [Google Scholar]
- Nagata, S.; Goldstein, P. The Fas death factor. Science 1995, 276, 1449–56. [Google Scholar]
- Chinnaiyan, A. M.; O’Rourke, K.; Lane, B. R.; Dixit, V. M. Interaction of CED-4 with CED-3 and CED-9, a molecular framework for cell death. Science 1997, 275, 1122–6. [Google Scholar]
- Akbar, A. N.; Salmon, M.; Savill, J.; Janossy, G. A possible role for bcl-2 in regulating T cell memory- a ‘balancing act’ between cell death and survival. Immunol Today 1993, 14, 526–31. [Google Scholar]
- Dianzani, U.; Bragardo, M.; DiFranco, D.; Alliaudi, C.; Scagni, P.; Buonfiglio, D.; Redoglia, V.; Bonissoni, S.; Correra, A.; Dianzani, I.; Ramenghi, U. Deficiency of the Fas Apoptosis Pathway Without Fas Gene Mutations in Pediatric Patients With Autoimmunity/Lymphoproliferation. Blood 1997, 89(8), 2871–2879. [Google Scholar]
- Maloy, K. J.; Powrie, F. Regulatory T cells in the control of immune pathology. Nat Immunol 2001, 2, 16–22. [Google Scholar]
- Ansari, M. J. I.; Salama, A. D.; Chitnis, T.; Smith, R. N.; Yagita, H.; Akiba, H.; Yamazaki, T.; Azuma, M.; Iwai, H.; Khoury, S. J.; Auchincloss, H., Jr.; Sayegh, M H. The programmed Death-1 (PD-1) Pathway Regulates Autoimmune Diabetes in Nonobese Diabetic (NOD) Mice. J. Exp Med 2003, 198(1), 63–69. [Google Scholar]
- Nishimura, H.; Okazaki, T.; Tanaka, Y.; Nakatani, K.; Hara, M.; Matsumori, A.; Sasayama, S.; Mizoguchi, A.; Hiai, H.; Minato, N.; Honjo, T. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science 2001, 291, 319–322. [Google Scholar]Mustelin, T.; Burn, P. Regulation of src family tyrosine kinases in lymphocytes. Trends in Biochemical Sciences 1993, 18, 215–220. [Google Scholar]
- Mustelin, T.; Burn, P. Regulation of src family tyrosine kinases in lymphocytes. Trends in Biochemical Sciences 1993, 18, 215–220. [Google Scholar]
- Cifone, M. G.; De Maria, R.; Roncaioli, P.; Rippo, M. R.; Azuma, M.; Lanier, L. L.; Santoni, A.; Testi, R. Apoptotic signaling through CD95 (Fas/APO-1) activates an acidic sphingomyelinase. J Exp Med 1993, 177, 1547–53. [Google Scholar]
- Chan, H. M. Metal accumulation and detoxification in humans. In Metal Metabolism in Aquatic Environments; Langston, W. J., Bebianno, M. J., Eds.; London; Chapman and Hall, 1998; pp. 415–438. [Google Scholar]
- Marsh, D. O.; Clarkson, T. W.; Myers, G. J.; Davidson, P. W.; Cox, C.; Cermichiari, E.; Tanner, M. A.; Lednar, W.; Shamlaye, C.; Choisy, O. The Seychelles study of fetal methylmercury exposure and child development: Introduction. Neurotoxicol 1995, 16, 583–596. [Google Scholar]
- Myers, G. J.; Davidson, P. W.; Cox, A.; Shamlaye, C. F.; Tanner, M. A.; Choisy, O.; Sloane-Reeves, J.; Marsh, D.; Cernichiari, E.; Choi, A. Neurodevelopmental outcomes of Seychellois children sixty-six months after in utero exposure to Methylmercury from a maternal fish diet: pilot study. Neurotoxicol 1995, 16(4), 639–652. [Google Scholar]
- Tchounwou, P. B.; Ayensu, W. K.; Ninashvili, N.; Sutton, D. Environmental Exposure to Mercury and its Toxico-Pathologic Implications to Public Health. Int. J. Environ. Res. Public Health 2004, 1, 39–73. [Google Scholar]
- ATSDR, Toxicological Profile for Mercury: TP-93/10. In Agency for Toxic Substances and Disease Registry; Centers for Disease Control: Atlanta, Georgia, 1999.
- Murphy, P. M.; Baggiolini, M.; Charo, I. F.; Hébert, C. A.; Horuk, R.; Matsushima, K.; Miller, L. H.; Oppenheim, J. J.; Power, C. A. International Union of Pharmacology. XXII. Nomenclature for Chemokine Receptors. Pharmacological Rev 2000, 52(1), 145–176. [Google Scholar]
- Gupta, S. K.; Gallego, C.; Johnson, G. L. Mitogenic pathways regulated by G proteins oncogenes. Molecular Biology and Cell 1992, 3, 123–128. [Google Scholar]
- Weiss, A.; Schlessinger, J. Switching signals on or off by receptor dimerization. Cell 1998, 94, 277–280. [Google Scholar]
- Steinberg, A. D.; Raveché, E. S.; Laskin, C. A.; Smith, H. R.; Santoro, T.; Miller, M. L.; Plotz, P. H. NIH conference. Systemic lupus erythematosus: insights from animal models. Ann Intern Med 1984, 100, 714–727. [Google Scholar]
- Drappa, J.; Kamen, L. A.; Chan, E.; Georgiev, M.; Ashany, D.; Marti, F.; King, P. D. Impaired T cell death and lupus-like autoimmunity in T cell-specific adapter protein-deficient mice. J. Exp Med 2003, 198(5), 809–821. [Google Scholar]
- Bishop, J. M. Molecular themes in oncogenes. Cell 1991, 64, 235–248. [Google Scholar]
- Hauptmann, R.; Swetly, P. A novel class of human type 1 interferons. Nuclei Acids Res 1985, 13, 4739–4749. [Google Scholar]
- Preble, O. T.; Rothko, K.; Klippel, J. H.; Friedman, R. M.; Johnston, M. I. Interferon-induced 2′–5′ adenylate synthetase in vitro by lymphocytes from systemic lupus erythematosus patients with and without circulating interferon. J. Exp. Med 1983, 157, 2140–2146. [Google Scholar]
- Adolf, G. R. Monoclonal antibodies and enzyme immunoassays specific for human interferon IFNω1: evidence that IFN-ω is a component of human leukocyte IFN. Virology 1990, 175, 410–417. [Google Scholar]
- Cederblad, B.; Blomberg, S.; Vallin, H.; et al. Patients with systemic lupus erythematosus have reduced numbers for circulating natural interferon-alpha-producing cells. J. Autoimmunol 1998, 11, 465–470. [Google Scholar]
- Blomberg, S.; Eloranta, M. L.; Cederblad, B.; et al. Presence of cutaneous interferon-α producing cells in patients with systemic lupus erythematosus. Lupus 2001, 10484–490. [Google Scholar]
- Hylton, W.; Cayley, J.; Dore, C.; Denman, A. M. 2′ 5′Oligoadenylate synthetase induction in lymphocytes of patients with connective tissue diseases. Ann. Rheum. Dis 1986, 45, 220–224. [Google Scholar]
- Ronnblom, L. E.; Alm, G. V.; Oberg, K. E. Autoimmunity after alpha-interferon therapy for malignant carcinoid tumor. Ann. Intern. Med 1991, 115, 178–183. [Google Scholar]
- Ytterberg, S. R.; Schnitzer, T. J. Serum interferon levels in patients with systemic lupus erythematosus. Arthritis Rheum 1982, 25, 401–406. [Google Scholar]
- Ronnblom, L. E.; Alm, G. V.; Oberg, K. E. Possible induction of systemic lupus erythematosus by interferon-α treatment in-patient with a malignant carcinoid tumor. J. Intern. Med 1990, 227, 207–210. [Google Scholar]
- Marrack, P.; Kappler, J.; Mitchell, T. Type I interferon keep activated T cells alive. J. Exp. Med 1999, 189, 521–529. [Google Scholar]
- Su, L.; David, M. Inhibition of B cell receptor-mediated apoptosis by IFN. J. Immunol 1999, 162, 6317–6321. [Google Scholar]
- Blanco, P.; Palucka, A. K.; Gill, M.; Pascual, V.; Banchereau, J. Induction of dendritic cell differentiation by IFN-α in systemic lupus erythymatosus. Science 2001, 294, 1540–1543. [Google Scholar]
- Ronnblom, L.; Alm, G. V. An etiopathogenic role for the type I IFN system in SLE. Trends Immunol 2001, 22, 427–431. [Google Scholar]
- Neubauer, R. H.; Goldstein, L.; Rabin, H.; Stebbing, N. Stimulation of in vitro immunoglobulin production by interferon-α. J. Immunol 1985, 134, 299–303. [Google Scholar]
- Moore, P. A.; Belvedere, O.; Orr, A.; Bly, S. member of the tumor necrosis factor family and B lymphocyte stimulator. Science 1999, 285, 260–263. [Google Scholar]
- Hultman, P.; Lindh, U.; Horsted-Bindsev, P. Activation of the immune system and systemic immune-complex deposits in Brown Norway rats with dental amalgam restorations. J. Dent. Res 1998, 77, 1415–1425. [Google Scholar]
- Hultman, P.; Bell, L. J.; Enestrom, S.; Pollard, K. M. Murine susceptibility to mercury. I. Autoantibody profiles and systemic immune deposits in inbred, congenic and intra-H-2 recombinant strains. Clin. Immunol. Immuno-Pathol 1992, 65, 98–108. [Google Scholar]
- Hultman, P.; Bell, L. J.; Eneström, S.; Pollard, K. M. Murine susceptibility to mercury. II. Autoantibody profiles and renal immune deposits in hybrid, backcross, and H-2d congenic mice. Clin. Immunol. Immuno. Pathol 1993, 68, 9–17. [Google Scholar]
- Kono, D. H.; Balomenos, D.; Pearson, D. L.; Park, M. S.; Gildebrandt, B.; Hultman, P.; Pollard, K. M. The prototypic Th2 autoimmunity induced by mercury is dependent on IFN- γ and not TH1/TH2 imbalance. J. Immunol 1998, 161, 234–240. [Google Scholar]
- Hanley, G. A.; Schiffenbauer, J.; Sobel, E. S. Class II haplotype differentially regulates immune response in HgCl2-treated mice. Clin Immunol Immunopathol 1997, 84, 328–333. [Google Scholar]
- Hanley, G. A.; Schiffenbauer, J.; Sobel, E. S. Resistance to HgCl2-induced autoimmunity in haplotype-heterozygous mice is an intrinsic property of B cells. J. Immunol 2002, 161(4), 1778–1789. [Google Scholar]
- Hirsch, F.; Kuhn, J.; Ventura, M.; Vial, M-C.; Fournie, G.; Druet, P. Autoimmunity induced by HgCl2 in Brown-Norway rats I. Production of monoclonal antibodies. J. Immunol 1986, 36, 3272–3276. [Google Scholar]
- Van der Pouw Kraan, T. C. T. M.; Van Gaalen, F. A.; Huizinga, T. W.; Pieterman, E.; Breedveld, F. C.; Verweij, C. L. Discovery of distinctive gene expression profiles in rheumatoid synovium using cDNA microarray technology: evidence for the existence of multiple pathways of tissue destruction and repair. Genes and Immunity 2003, 4, 187–196. [Google Scholar]
- Fukui, Y.; Hashimoto, O.; Sanui, T.; Oono, T.; Koga, H.; Abe, M.; Inayoshi, A.; Noda, M.; Oike, M.; Shirai, T.; Sasazuki, T. Haematopoietic cell specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature 2001, 412, 826–831. [Google Scholar]
- Firestein, G. S.; Manning, A. M. Signal transduction and transcription factors in rheumatic disease. Arthritis Rheum 1999, 42, 609–21. [Google Scholar]
- Shi, X.; Xie, C.; Kreska, D.; Richardson, J. A.; Mohan, C. Genetic Dissection of SLE. SLE1 and FAS Impact Alternate Pathways Leading to Lymphoproliferative Autoimmunity. J. Exp. Med 2002, 196, 281–292. [Google Scholar]
- Manoury-Schwartz, B.; Chiocchia, G.; Bessis, N.; Abehsira-Amar, O.; Batteux, F.; Muller, S.; Huang, S.; Boissier, M. C.; Fournier, C. High susceptibility to collagen-induced arthritis in mice lacking IFN- γ receptors. J. Immunol 1997, 158, 5501–5506. [Google Scholar]
- Vermeire, K.; Heremans, H.; Vandeputte, M.; Huang, S.; Billiau, A.; Matthys, P. Accelerated collagen-induced arthritis in IFN-γ receptor-deficient mice. J. Immunol 1997, 158, 5507–5513. [Google Scholar]
- Shan, X. C.; Bourdeau, A.; Rhoton, A. Characterization and mapping to human chromosome 8q24.3 of Ly-6-related gene 9804 encoding an apparent homologue of mouse TSA-1. J. Immunol 1998, 160, 197–208. [Google Scholar]
- Andre, I.; Gonzalez, A.; Wang, B.; Katz, J.; Benoist, C.; Mathis, D. Checkpoints in the progression of autoimmune disease: lessons from diabetes models. Proc Natl Acad Sci USA 1996, 93, 2260–2274. [Google Scholar]
- Hultgren, B.; Huang, X.; Dybdal, N.; Stewart, T. A. Genetic absence of gamma-interferon delays but does not prevent diabetes in NOD mice. Diabetes 1996, 45, 812–819. [Google Scholar]
- Hall, A. Ras-related proteins. Current Opinion in Cellular Biology 1993, 5, 265–268. [Google Scholar]
- Lowy, D. R.; Williamson, B. M. Function and regulation of Ras. Annual Review of Biochemistry 1993, 62, 851–891. [Google Scholar]
- Ayensu, W. K.; Tchounwou, P. B.; McMurray, R. W. Molecular and Cellular Mechanisms Associated with Autoimmune Diseases. Int. J. Environ. Res. Public Health 2004, 1, 39–74. [Google Scholar]
- Affymetrix manual: Affymetrix Technical Gene Chip manual version 5.0 2002.
- Tsao, B. P.; Cantor, R. M.; Kalunian, K. C.; et al. Evidence for linkage of a candidate chromosome 1 region to human systemic lupus erythematosus. J. Clin Invest 1997, 99, 725–731. [Google Scholar]
- Drysdale, C. M.; McGraw, D. W.; Stack, C. B.; et al. Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci USA 2000, 97, 10483–8. [Google Scholar]
- Ruiz-Vera, A.; Gonzα1lez De Buitrago, G.; Martinez-A, C. Implication of calpain in caspase activation during B cell clonal deletion. EMBO J 1999, 18, 4988–4998. [Google Scholar]
- Wu, M.; Arsula, M.; Bellas, R. E.; FitzGerald, M. J.; Lee, H.; Schauer, S. L.; Sherr, D. H.; Sonensheink, G. E. Inhibition of NF-κB/Rel induces apoptosis of murine B cells. Mol. Cell Biol 1996, 16, 5015–5025. [Google Scholar]
- Massagué, J.; Wotton, D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J 2000, 19, 1745–1754. [Google Scholar]
- ten Dijke, P.; Miyazono, K.; Heldin, C-H. Signalling input converge on nuclear effectors in TGF-β signaling. Trends Biochem. Sci 2000, 25, 64–70. [Google Scholar]
- Zhang, Y.; Derynck, R. Regulation of Smad signalling by protein associations and signalling crosstalk. Trends Cell Biol 1999, 9, 274–279. [Google Scholar]
- Wakeland, E. K.; Liu, K.; Graham, R. R.; Behrens, T. W. Delineating the genetic basis of systemic lupus erythematosus. Immunity 2001, 15, 397–408. [Google Scholar]
- Moser, L. M.; Neas, B. R.; Salmon, J. E.; Yu, H.; Gray-McGuire, C.; Asundi, N.; Fox, J.; Kelly, J.; Henshall, S.; Bacino, D.; Dietz, M.; Hogue, R.; Koelsch, G.; Nightingale, L.; Shaver, T.; Abdou, N. I.; Albert, D. A.; Carson, C.; Petr, M.; Treadwell, E. L.; James, J. A.; Harley, J. B. Genome scan of human systemic lupus erythematosus: Evidence for linkage on chromosome 1q in African-American pedigrees. Proc. Nat. Acad. Sci. USA 1998, 95, 14869–74. [Google Scholar]
- Deapen, D. M.; Escalante, A.; Weinrib, L.; Horwitz, D. A.; Mack, M. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum 1992, 35, 311–318. [Google Scholar]
- Gaffney, P. M.; Ortmann, W. A.; Selby, S. A.; et al. Genome screening in human systemic lupus erythematosus: results from a second Minnesota cohort and combined analyses of sib-pair families. Am J Human Genet 2000, 66, 547–556. [Google Scholar]
© 2006 MDPI. All rights reserved.
Share and Cite
Ayensu, W.K.; Tchounwou, P.B. Microarray Analysis of Mercury-Induced Changes in Gene Expression in Human Liver Carcinoma (HepG2) Cells: Importance in Immune Responses. Int. J. Environ. Res. Public Health 2006, 3, 141-173. https://doi.org/10.3390/ijerph2006030018
Ayensu WK, Tchounwou PB. Microarray Analysis of Mercury-Induced Changes in Gene Expression in Human Liver Carcinoma (HepG2) Cells: Importance in Immune Responses. International Journal of Environmental Research and Public Health. 2006; 3(2):141-173. https://doi.org/10.3390/ijerph2006030018
Chicago/Turabian StyleAyensu, Wellington K., and Paul B. Tchounwou. 2006. "Microarray Analysis of Mercury-Induced Changes in Gene Expression in Human Liver Carcinoma (HepG2) Cells: Importance in Immune Responses" International Journal of Environmental Research and Public Health 3, no. 2: 141-173. https://doi.org/10.3390/ijerph2006030018





