Microbial Arsenic Methylation in Soil and Uptake and Metabolism of Methylated Arsenic in Plants: A Review
Abstract
:1. Introduction
2. Arsenic Methylation
2.1. Biochemistry Process of Arsenic Methylation
2.2. Molecular Mechanisms of Arsenic Methylation
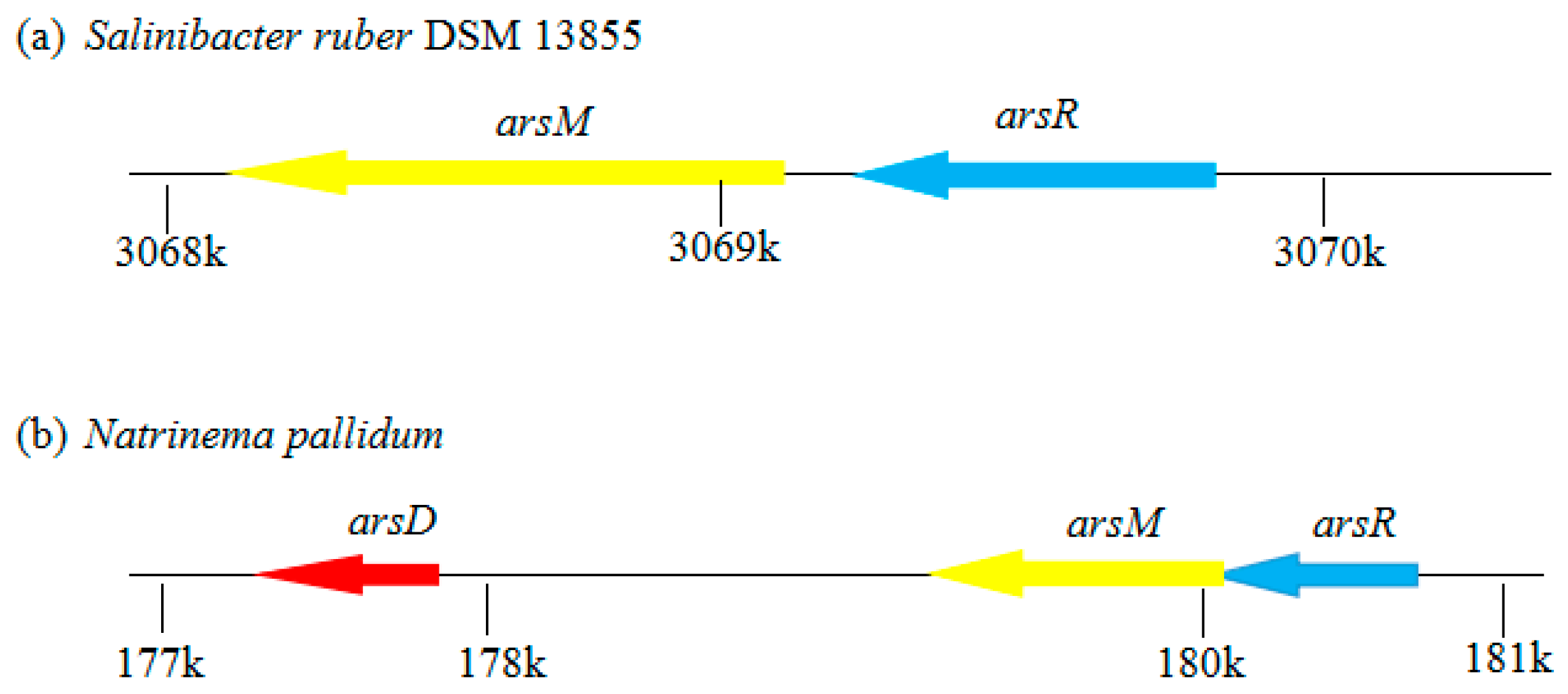
2.3. Gene Characterization of Arsenic Methylation
2.4. Factors Affecting Microbial Arsenic Methylation in Soil
2.5. Uptake and Metabolism of Methylated Arsenic in Plants
2.5.1. Effect of Arsenic Methylation in Soil on Absorption and Metabolism of Plant Methylated Arsenic
2.5.2. Effect of Arsenic Methylation in Soil on Remediation of Arsenic Pollution
2.5.3. The Toxicity of Methylated Arsenic on Organisms
2.6. Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bhattacharya, P.; Welch, A.H.; Stollenwerk, K.G.; McLaughlin, M.J.; Bundschuh, J.; Panaullah, G. Arsenic in the environment: Biology and Chemistry. Sci. Total Environ. 2007, 379, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Brammer, H.; Ravenscroft, P. Arsenic in groundwater: A threat to sustainable agriculture in South and South-East Asia. Environ. Int. 2009, 35, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Lado, L.; Sun, G.F.; Berg, M. Groundwater arsenic contamination throughout China. Science 2013, 341, 866–868. [Google Scholar] [CrossRef] [PubMed]
- Akter, K.F.; Owens, G.; Davey, D.E.; Naidu, R. Arsenic speciation and toxicity in biological systems. Rev. Environ. Contam. Toxicol. 2005, 184, 97–149. [Google Scholar]
- Huang, J.H.; Hu, K.N.; Decker, B. Organic arsenic in the soil environment: Speciation, occurrence, transformation, and adsorption behavior. Water Air Soil Pollut. 2011, 219, 401–415. [Google Scholar] [CrossRef]
- Meharg, A.A.; Williams, P.N.; Adomako, E.; Lawgali, Y.Y.; Deacon, C.; Villada, A.; Cambell, R.C.J.; Sun, G.X.; Zhu, Y.G.; Feldmann, J.; et al. Geographical variation in total and inorganic arsenic content of polished (white) rice. Environ. Sci. Technol. 2009, 43, 1612–1617. [Google Scholar] [CrossRef]
- Arao, T.; Kawasaki, A.; Baba, K.; Matsumoto, S. Effects of Arsenic Compound Amendment on Arsenic Speciation in Rice Grain. Environ. Sci. Technol. 2011, 45, 1291–1297. [Google Scholar] [CrossRef]
- Zheng, M.Z.; Cai, C.; Hu, Y.; Sun, G.X.; Williams, P.N.; Cui, H.J.; Li, G.; Zhao, F.J.; Zhu, Y.G. Spatial distribution of arsenic and temporal variation of its concentration in rice. New Phytol. 2011, 189, 200–209. [Google Scholar] [CrossRef]
- Lomax, C.; Liu, W.J.; Wu, L.Y.; Xue, K.; Xiong, J.B.; Zhou, J.Z.; Mcgrath, S.P.; Meharg, A.A.; Miller, A.J.; Zhao, F.J. Methylated arsenic species in plants originate from soil microorganisms. New Phytol. 2012, 193, 665–672. [Google Scholar] [CrossRef]
- Ackerman, A.H.; Creed, P.A.; Parks, A.N.; Fricke, M.W.; Schwegel, C.A.; Creed, J.T.; Heitkemper, D.T.; Velal, N.P. Comparison of a Chemical and Enzymatic Extraction of Arsenic from Rice and an Assessment of the Arsenic Absorption from Contaminated Water by Cooked Rice. Environ. Sci. Technol. 2005, 39, 5241–5246. [Google Scholar] [CrossRef]
- Naranmandura, H.; Ibata, K.; Suzuki, K.T. Toxicity of dimethylmonothioarsinic acid toward human epidermoid carcinoma A431 cells. Chem. Res. Toxicol. 2007, 20, 1120–1125. [Google Scholar] [CrossRef] [PubMed]
- Kerl, C.F.; Schindele, R.A.; Brüggenwirth, L.; Colina Blanco, A.E.; Rafferty, C.; Clemens, S.; Planer-Friedrich, B. Methylated thioarsenates and monothioarsenate differ in uptake, transformation, and contribution to total arsenic translocation in rice plants. Environ. Sci. Technol. 2019, 53, 5787–5796. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, R.; Shao, K.; Thomas, D.J.; Sams, R.; Cowden, J. AS3MT, GSTO, and PNP polymorphisms: Impact on arsenic methylation and implications for disease susceptibility. Environ. Res. 2014, 132, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Savage, L.; Carey, M.P.; Williams, P.N.; Meharg, A.A. Maritime deposition of organic and inorganic arsenic. Environ. Sci. Technol. 2019, 53, 7288–7295. [Google Scholar] [CrossRef] [PubMed]
- Savage, L.; Carey, M.; Hossain, M.; Islam, M.R.; de Silva, P.M.C.; Williams, P.N.; Meharg, A.A. Elevated trimethylarsine oxide and inorganic arsenic in northern hemisphere summer monsoonal wet deposition. Environ. Sci. Technol. 2017, 51, 12210–12218. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Xue, X.M.; Kappler, A.; Rosen, B.P.; Meharg, A.A. Linking Genes to Microbial Biogeochemical Cycling: Lessons from Arsenic. Environ. Sci. Technol. 2017, 51, 7326–7339. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.J.; Waters, S.B.; Styblo, M. Elucidating the pathway for arsenic methylation. Toxicol. Appl. Pharm. 2004, 198, 319–326. [Google Scholar] [CrossRef]
- Huang, H.; Jia, Y.; Sun, G.; Zhu, Y.G. Arsenic speciation and volatilization from flooded paddy soils amended with different organic matters. Environ. Sci. Technol. 2012, 46, 2163–2168. [Google Scholar] [CrossRef]
- Le, X.C.; Lu, X.; Ma, M.; Cullen, W.R.; Aposhian, H.V.; Zheng, B. Speciation of key arsenic metabolic intermediates in human urine. Anal. Chem. 2000, 72, 5172–5177. [Google Scholar] [CrossRef]
- Muñoz, L.; Meneses, M.; Pismante, P.; Andonie, O.; Queirolo, F.; Stegen, S. Methodological validation for the determination of toxic arsenic species in human urine using HPLC with ICP-MS. J. Chil. Chem. Soc. 2014, 59, 2432–2436. [Google Scholar]
- Turpeinen, R.; Pantsar-Kallio, M.; Kairesalo, T. Role of microbes in controlling the speciation of arsenic and production of arsines in contaminated soils. Sci. Total Environ. 2002, 285, 133–145. [Google Scholar] [CrossRef]
- Mestrot, A.; Feldmann, J.; Krupp, E.M.; Hossain, M.S.; Roman-Ross, G. Field Fluxes and Speciation of Arsines Emanating from Soils. Environ. Sci. Technol. 2011, 45, 1798–1804. [Google Scholar] [CrossRef] [PubMed]
- Savage, L.; Carey, M.; Williams, P.N.; Meharg, A.A. Biovolatilization of arsenic as arsines from seawater. Environ. Sci. Technol. 2018, 52, 3968–3974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Challenger, F.; Higginbottom, C. The production of trimethylarsine by Penicillium brevicaule (Scopulariopsis brevicaulis). Biochem. J. 1935, 29, 1757–1778. [Google Scholar] [CrossRef] [Green Version]
- Challenger, F. Biological Methylation. Chem. Rev. 1945, 36, 315–361. [Google Scholar] [CrossRef]
- Qin, J.; Rosen, B.P.; Zhang, Y.; Wang, G.J.; Franke, S.; Rensing, C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 2075–2080. [Google Scholar] [CrossRef] [Green Version]
- Bentley, R.; Chasteen, T.G. Microbial methylation of metalloids: Arsenic, antimony, and bismuth. Microbiol. Mol. Biol. Rev. 2002, 66, 250–271. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.P.; Sun, G.X.; Jia, Y.; Meharg, A.A.; Zhu, Y.G. A review on completing arsenic biogeochemical cycle: Microbial volatilization of arsines in environment. J. Environ. Sci. 2014, 26, 371–381. [Google Scholar] [CrossRef]
- Li, J.L.; Sun, Y.Q.; Zhang, X.; Hu, Y.J.; Li, T.; Zhang, X.M.; Wang, Z.; Wu, S.L.; Wu, Z.X.; Chen, B.D. A methyltransferase gene from arbuscular mycorrhizal fungi involved in arsenic methylation and volatilization. Chemosphere 2018, 209, 392–400. [Google Scholar] [CrossRef]
- Verma, S.; Verma, P.K.; Chakrabarty, D. Arsenic Bio-volatilization by Engineered Yeast Promotes Rice Growth and Reduces Arsenic Accumulation in Grains. Int. J. Environ. Res. 2019, 13, 475–485. [Google Scholar] [CrossRef]
- Yin, X.X.; Wang, L.H.; Zhang, Z.C.; Fan, G.L.; Liu, J.J.; Sun, K.Z.; Sun, G.X. Biomethylation and volatilization of arsenic by model protozoan Tetrahymena pyriformis under different phosphate regimes. Int. J. Environ. Res. Public Health 2017, 14, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakharyan, R.A.; Aposhian, H.V. Arsenite Methylation by Methylvitamin B12and Glutathione Does Not Require an Enzyme* 1,* 2,* 3. Toxicol. Appl. Pharmacol. 1999, 154, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, T.; Kobayashi, Y.; Cui, X.; Hirano, S. A new metabolic pathway of arsenite: Arsenic–glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch. Toxicol. 2005, 79, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Lehr, C.R.; Polishchuk, E.; Radoja, U.; Cullen, W.R. Demethylation of methylarsenic species by Mycobacterium neoaurum. Appl. Organomet. Chem. 2003, 17, 831–834. [Google Scholar] [CrossRef]
- Yoshinaga, M.; Cai, Y.; Rosen, B.P. Demethylation of methylarsonic acid by a microbial community. Environ. Microbiol. 2011, 13, 1205–1215. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Ye, J.; Xue, X.M.; Zhu, Y.G. Arsenic demethylation by a C·As lyase in cyanobacterium Nostoc sp. PCC 7120. Environ. Sci. Technol. 2015, 49, 14350–14358. [Google Scholar] [CrossRef]
- Ye, J.; Rensing, C.; Rosen, B.P.; Zhu, Y.G. Arsenic biomethylation by photosynthetic organisms. Trends Plant Sci. 2012, 17, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Huang, H.; Zhong, M.; Wang, F.H.; Zhang, L.M.; Zhu, Y.G. Microbial arsenic methylation in soil and rice rhizosphere. Environ. Sci. Technol. 2013, 47, 3141–3148. [Google Scholar] [CrossRef]
- Chen, J.; Sun, G.X.; Wang, X.X.; de Lorenzo, V.; Rosen, B.P.; Zhu, Y.G. Volatilization of arsenic from polluted soil by Pseudomonas putida engineered for expression of the arsM arsenic (III) S-adenosine methyltransferase gene. Environ. Sci. Technol. 2014, 48, 10337–10344. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Cao, T.T.; Tang, Z.; Shen, Q.R.; Rosen, B.P.; Zhao, F.J. Arsenic Methylation and Volatilization by Arsenite S-Adenosylmethionine Methyltransferase in Pseudomonas alcaligenes NBRC14159. Appl. Environ. Microbiol. 2015, 81, 2852–2860. [Google Scholar] [CrossRef] [Green Version]
- Kuramata, M.; Sakakibara, F.; Kataoka, R.; Abe, T.; Asano, M.; Baba, K.; Takagi, K.; Ishikawa, S. Arsenic biotransformation by Streptomyces sp. isolated from rice rhizosphere. Environ. Microbiol. 2015, 17, 1897–1909. [Google Scholar] [CrossRef] [PubMed]
- Reid, M.C.; Maillard, J.; Bagnoud, A.; Falquet, L.; Le, V.P.; Bernier-Latmani, R. Arsenic Methylation Dynamics in a Rice Paddy Soil Anaerobic Enrichment Culture. Environ. Sci. Technol. 2017, 51, 10546–10554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.; Xu, Y.; Packianathan, C.; Gao, F.; Chen, C.; Zhang, J.; Shen, Q.R.; Rosen, B.P.; Zhao, F.J. Arsenic methylation by a novel ArsM As(III) S-adenosylmethionine methyltransferase that requires only two conserved cysteine residues. Mol. Microbiol. 2018, 107, 265–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, D.P.; Alexander, M. Production of trimethylarsine gas from various arsenic compounds by three sewage fungi. Bull. Environ. Contam. Toxicol. 1973, 9, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Cullen, W.R.; Reimer, K.J. Arsenic speciation in the environment. Chem. Rev. 1989, 89, 713–764. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.B.; Su, S.M.; Jiang, X.L.; Li, L.F.; Bai, L.Y.; Zhang, Y.R. Capability of Pentavalent Arsenic Bioaccumulation and Biovolatilization of Three Fungal Strains under Laboratory Conditions. Clean-Soil Air Water 2010, 38, 238–241. [Google Scholar] [CrossRef]
- Srivastava, P.K.; Vaish, A.; Dwivedi, S.; Chakrabarty, D.; Singh, N.; Tripathi, R.D. Biological removal of arsenic pollution by soil fungi. Sci. Total Environ. 2011, 409, 2430–2442. [Google Scholar] [CrossRef]
- Kavitha, M.; Jie, Q.; Rosen, B.P. Identification of Catalytic Residues in the As(III) S-Adenosylmethionine Methyltransferase. Biochemistry 2012, 51, 944–951. [Google Scholar]
- Zhao, F.J.; Harris, E.; Yan, J.; Ma, J.C.; Zhu, Y.G. Arsenic methylation in soils and its relationship with microbial arsM abundance and diversity, and as speciation in rice. Environ. Sci. Technol. 2013, 47, 7147–7154. [Google Scholar] [CrossRef]
- Wang, Y.H.; Li, P.; Jiang, Z.; Liu, H.; Wei, D.Z.; Wang, H.L.; Wang, Y.X. Diversity and abundance of arsenic methylating microorganisms in high arsenic groundwater from Hetao Plain of Inner Mongolia, China. Ecotoxicology 2018, 27, 1047–1057. [Google Scholar] [CrossRef]
- Chen, J.; Qin, J.; Zhu, Y.G.; de Lorenzo, V.; Rosen, B.P. Engineering the soil bacterium Pseudomonas putida for arsenic methylation. Appl. Environ. Microbiol. 2013, 79, 4493–4495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, J.; Chang, Y.; Yan, Y.; Xiong, J.; Xue, X.M.; Yuan, D.X.; Sun, G.X.; Zhu, Y.G.; Miao, W. Identification and characterization of the arsenite methyltransferase from a protozoan, Tetrahymena pyriformis. Aquat. Toxicol. 2014, 149, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Chen, C.; Zhang, J.; Tang, Z.; Shen, Q.R.; Rosen, B.P.; Zhao, F.J. Efficient Arsenic Methylation and Volatilization Mediated by a Novel Bacterium from an Arsenic-Contaminated Paddy Soil. Environ. Sci. Technol. 2016, 50, 6389–6396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, S.; Verma, P.K.; Meher, A.K.; Bansiwal, A.K.; Tripathi, R.D.; Chakrabarty, D. A novel fungal arsenic methyltransferase, WaarsM reduces grain arsenic accumulation in transgenic rice (Oryza sativa L.). J. Hazard. Mater. 2018, 344, 626–634. [Google Scholar] [CrossRef]
- Vriens, B.; Lenz, M.; Charlet, L.; Berg, M.; Winkel, L. Natural wetland emissions of methylated trace elements. Nat. Commun. 2014, 5, 30–35. [Google Scholar] [CrossRef] [Green Version]
- Ascar, L.; Ahumada, I.; Richter, P. Influence of redox potential (Eh) on the availability of arsenic species in soils and soils amended with biosolid. Chemosphere 2008, 72, 1548–1552. [Google Scholar] [CrossRef]
- Zecchin, S.; Corsini, A.; Martin, M.; Cavalca, L. Influence of water management on the active root-associated microbiota involved in arsenic, iron, and sulfur cycles in rice paddies. Appl. Microbiol. Biotechnol. 2017, 101, 6725–6738. [Google Scholar] [CrossRef]
- Chen, C.; Huang, K.; Xie, W.Y.; Chen, S.H.; Tang, Z.; Zhao, F.J. Microbial processes mediating the evolution of methylarsine gases from dimethylarsenate in paddy soils. Environ. Sci. Technol. 2017, 51, 13190–13198. [Google Scholar] [CrossRef]
- Zhao, F.J.; Ma, J.F.; Meharg, A.A.; Mcgrath, S.P. Arsenic uptake and metabolism in plants. New Phytol. 2009, 181, 777–794. [Google Scholar] [CrossRef]
- Li, R.Y.; Ago, Y.; Liu, W.J.; Mitani, N.; Feldmann, J.; Mcgrath, S.P.; Ma, J.F.; Zhao, F.J. The rice aquaporin Lsi1 mediates uptake of methylated arsenic species. Plant Physiol. 2009, 150, 2071–2080. [Google Scholar] [CrossRef] [Green Version]
- Raab, A.; Williams, P.N.; Meharg, A.A.; Feldmann, J. Uptake and translocation of inorganic and methylated arsenic species by plants. Environ. Chem. 2007, 4, 197–203. [Google Scholar] [CrossRef] [Green Version]
- Abedin, M.J.; Feldmann, J.; Meharg, A.A. Uptake kinetics of arsenic species in rice plants. Plant Physiol. 2002, 128, 1120–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carey, A.M.; Norton, G.J.; Deacon, C.; Scheckel, K.G.; Lombi, E.; Punshon, T.; Guerinot, M.L.; Lanzirotti, A.; Newville, M.; Choi, Y.; et al. Phloem transport of arsenic species from flag leaf to grain during grain filling. New Phytol. 2011, 192, 87–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.; Li, J.; Wang, H.Y.; Zheng, R.L.; Sun, G.X. Evaluation of bioaugmentation and bio-stimulation on arsenic remediation in soil through biovolatilization. Environ. Sci. Pollut. Res. 2017, 24, 21739–21749. [Google Scholar] [CrossRef]
- Huang, K.; Chen, C.; Shen, Q.R.; Rosen, B.P.; Zhao, F.J. Genetically engineering Bacillus subtilis with a heat-resistant arsenite methyltransferase for bioremediation of arsenic-contaminated organic waste. Int. J. Environ. Res. Public Health 2015, 81, 6718–6724. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xu, Y.; Cao, T.T.; Chen, J.; Rosen, B.P.; Zhao, F.J. Arsenic methylation by a genetically engineered Rhizobium-legume symbiont. Plant Soil 2017, 416, 259–269. [Google Scholar] [CrossRef]
- Jia, Y.; Huang, H.; Sun, G.X.; Zhao, F.J.; Zhu, Y.G. Pathways and relative contributions to arsenic volatilization from rice plants and paddy soil. Environ. Sci. Technol. 2012, 46, 8090–8096. [Google Scholar] [CrossRef]
- Meng, X.Y.; Qin, J.; Wang, L.H.; Duan, G.L.; Sun, G.X.; Wu, H.L.; Chu, C.C.; Ling, H.Q.; Rosen, B.P.; Zhu, Y.G. Arsenic biotransformation and volatilization in transgenic rice. New Phytol. 2011, 191, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Urík, M.; Čerňanský, S.; Ševc, J.; Šimonovičová, A.; Littera, P. Biovolatilization of arsenic by different fungal strains. Water Air Soil Pollut. 2007, 186, 337–342. [Google Scholar] [CrossRef]
- Styblo, M.; Del Razo, L.M.; Vega, L.; Germolec, D.R.; LeCluyse, E.L.; Hamilton, G.A.; Reed, W.; Wang, C.Q.; Cullen, W.R.; Thomas, D.J. Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells. Arch. Toxicol. 2000, 74, 289–299. [Google Scholar] [CrossRef]
- Dopp, E.; Hartmann, L.M.; Florea, A.M.; Von Recklinghausen, U.; Pieper, R.; Shokouhi, B.; Rettenmeier, A.W.; Hirner, A.; Obe, G. Uptake of inorganic and organic derivatives of arsenic associated with induced cytotoxic and genotoxic effects in Chinese hamster ovary (CHO) cells. Toxicol. Appl. Pharmacol. 2004, 201, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Dopp, E.; von Recklinghausen, U.; Hippler, J.; Diaz-Bone, R.A.; Richard, J.; Zimmermann, U.; Rettenmeier, A.W.; Hirner, A.V.; Aschner, M. Toxicity of Volatile Methylated Species of Bismuth, Arsenic, Tin, and Mercury in Mammalian Cells In Vitro. J. Toxicol.-Toxin Rev. 2011, 2011, 503576. [Google Scholar]
- Rahman, M.A.; Hasegawa, H.; Rahman, M.; Miah, M.; Tasmin, A. Straight-head disease of rice (Oryza sativa L.) induced by arsenic toxicity. Environ. Exp. Bot. 2008, 62, 54–59. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.Z.; Li, G.; Sun, G.X.; Shim, H.; Cai, C. Differential toxicity and accumulation of inorganic and methylated arsenic in rice. Plant Soil 2013, 365, 227–238. [Google Scholar] [CrossRef]
- Duncan, E.G.; Maher, W.A.; Foster, S.D. Dimethylarsenate (DMA) exposure influences germination rates, arsenic uptake and arsenic species formation in wheat. Chemosphere 2017, 181, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.X.; Van de, W.T.; Alava, P.; Tack, F.; Du, L.G. Arsenic in cooked rice: Effect of chemical, enzymatic and microbial processes on bioaccessibility and speciation in the human gastrointestinal tract. Environ. Pollut. 2012, 162, 241–246. [Google Scholar] [CrossRef]



| Types | Main Methylated Arsenic Species | Major Origin | References |
|---|---|---|---|
| Soil | MMAs(V), DMAs(V), TMAs(V)O, TETRAs | Methylation mediated by microbes | [17,18] |
| Rice grain | DMAs(V), DMMTA | Methylation mediated by microbes in soil | [6,9,12] |
| Human | MMAs(V), DMAs(V) | Enzymatic methylation in vivo | [19,20] |
| Atmosphere | DMAs(III), TMAs(III), TMAs(V)O | Volatilization of methylated arsine | [21,22] |
| Ocean | TMAs(V)O | Atmospheric deposition | [14,15,23] |
| Strain | Major Product(s) | Source | Substrate | References |
|---|---|---|---|---|
| Bacteria | ||||
| Pseudomonas putida (with R. palustris arsM) | DMAs(V) | Soil | As(III), As(V) | [39] |
| Pseudomonas alcaligenes NBRC14159 | DMAs(V) | _ | As(III) | [40] |
| Streptomyces sp. | MMAs(V), DMAs(V) | Rice rhizosphere | As(III) | [41] |
| Sulfate-reducing bacteria | DMAs(V) | Mekong Delta paddy soil | As(III) | [42] |
| Bacillus strain | DMAs(V) | Manure compost | As(III), MMAs(III) | [43] |
| Fungi | ||||
| Scopulariopsis brevicaulis | TMAs(III) | _ | As(III) | [27] |
| Gliocladium roseum | TMAs(III) | Sewage | DMAs(V), MMAs(V) | [44] |
| Penicillium sp. | TMAs(III) | Sewage | DMAs(V), MMAs(V) | [44] |
| Aspergillus sydowi | TMAs(III) | _ | As(III), As(V) | [45] |
| Penicillium janthinellum | Total volatile arsenic | Soil | As(V) | [46] |
| Trichoderma asperellum | Total volatile arsenic | Soil | As(V) | [46] |
| Trichoderma sp. | Total volatile arsenic | Soil | As(V) | [47] |
| Aspergillus sp. | Total volatile arsenic | Soil | As(V) | [47] |
| Neocosmospora sp. | Total volatile arsenic | Soil | As(V) | [47] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di, X.; Beesley, L.; Zhang, Z.; Zhi, S.; Jia, Y.; Ding, Y. Microbial Arsenic Methylation in Soil and Uptake and Metabolism of Methylated Arsenic in Plants: A Review. Int. J. Environ. Res. Public Health 2019, 16, 5012. https://doi.org/10.3390/ijerph16245012
Di X, Beesley L, Zhang Z, Zhi S, Jia Y, Ding Y. Microbial Arsenic Methylation in Soil and Uptake and Metabolism of Methylated Arsenic in Plants: A Review. International Journal of Environmental Research and Public Health. 2019; 16(24):5012. https://doi.org/10.3390/ijerph16245012
Chicago/Turabian StyleDi, Xuerong, Luke Beesley, Zulin Zhang, Suli Zhi, Yan Jia, and Yongzhen Ding. 2019. "Microbial Arsenic Methylation in Soil and Uptake and Metabolism of Methylated Arsenic in Plants: A Review" International Journal of Environmental Research and Public Health 16, no. 24: 5012. https://doi.org/10.3390/ijerph16245012




