Modeling Schistosoma japonicum Infection under Pure Specification Bias: Impact of Environmental Drivers of Infection
Abstract
:1. Introduction
2. Materials and Methods
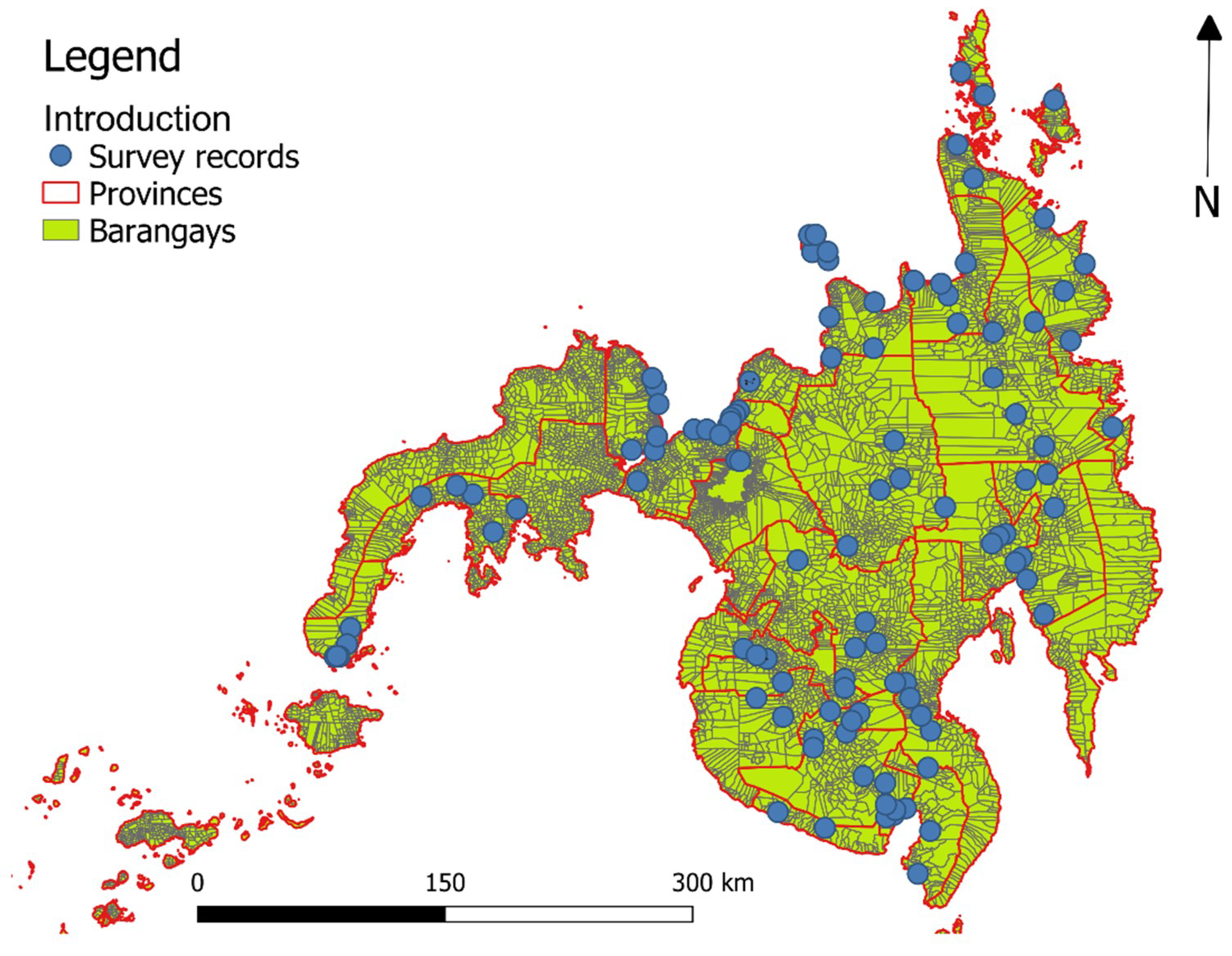
2.1. Data on Human Schistosoma japonicum Infection, Study Area, and Sampling Design
2.2. Environmental and Geographical Data
2.3. Convolution Model (Individual-Level Model)
2.4. Ecological Model (Group-Level Model)
2.5. Model Validation
2.6. Software and Data Sources
2.7. Ethics Approval
3. Results
3.1. Convolution Model
3.2. Ecological Model
3.3. Convolution Versus Ecological Model
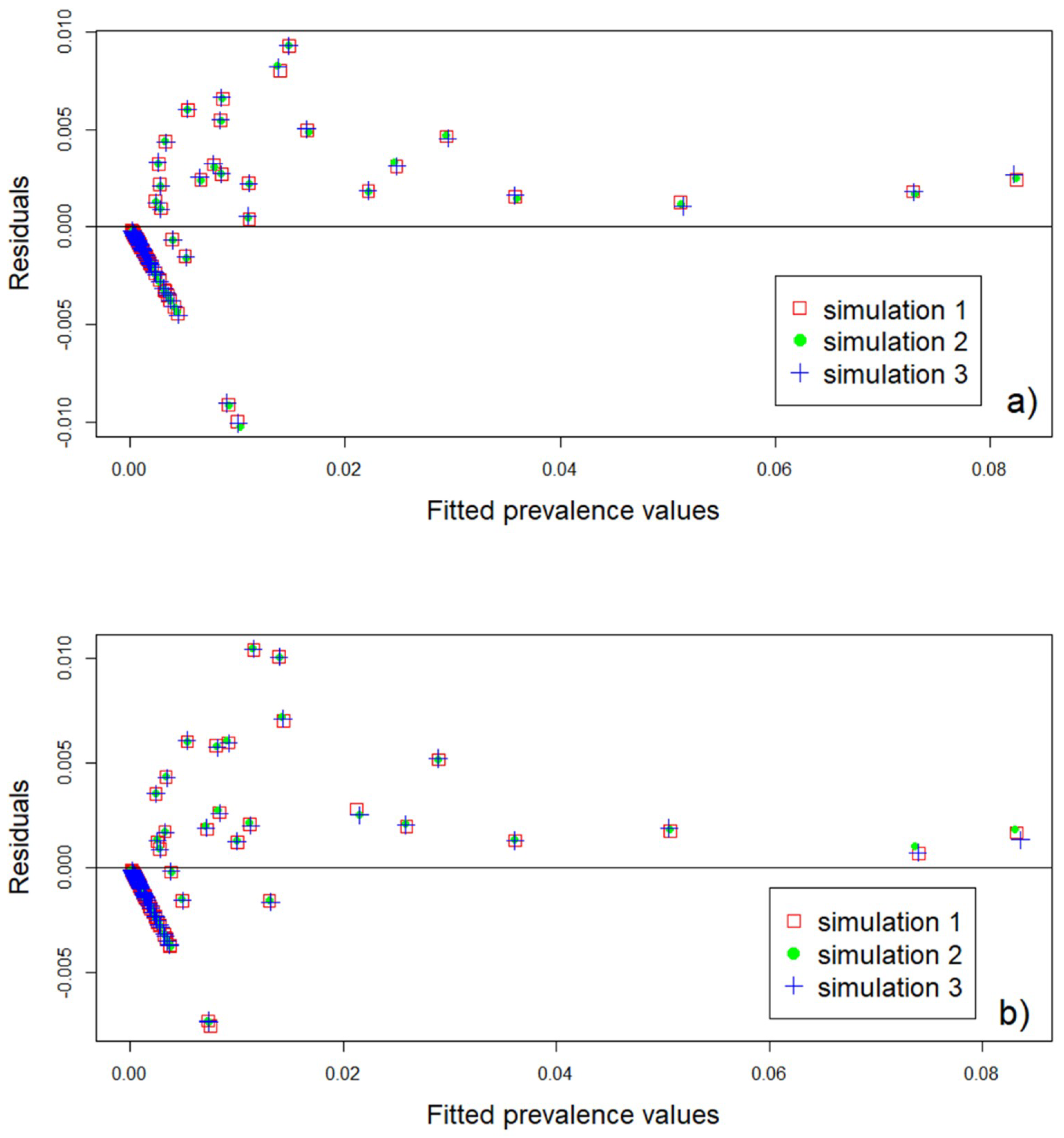
3.4. Model Validation
4. Discussion
Suggestions
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jia, T.W.; Zhou, X.N.; Wang, X.H.; Utzinger, J.; Steinmann, P.; Wu, X.H. Assessment of the age-specific disability weight of chronic schistosomiasis japonica. Bull. World Health Organ. 2007, 85, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Tarafder, M.R.; Balolong, E.; Carabin, H.; Belisle, P.; Tallo, V.; Joseph, L.; Alday, P.; Gonzales, R.O.; Riley, S.; Olveda, R.; et al. A cross-sectional study of the prevalence of intensity of infection with Schistosoma japonicum in 50 irrigated and rain-fed villages in Samar Province, the Philippines. BMC Public Health 2006, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Wang, X.H.; Yang, G.J.; Wu, X.H.; Qi, Y.L.; Li, H.J.; Zhou, X.N. An integrated approach to identify distribution of Oncomelania hupensis, the intermediate host of Schistosoma japonicum, in a mountainous region in China. Int. J. Parasitol. 2008, 38, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- King, C.H.; Dickman, K.; Tisch, D.J. Reassessment of the cost of chronic helmintic infection: A meta-analysis of disability-related outcomes in endemic schistosomiasis. Lancet 2005, 365, 1561–1569. [Google Scholar] [CrossRef]
- Walz, Y.; Wegmann, M.; Dech, S.; Vounatsou, P.; Poda, J.-N.; N’Goran, E.K.; Utzinger, J.; Raso, G. Modeling and Validation of Environmental Suitability for Schistosomiasis Transmission Using Remote Sensing. PLoS Negl. Trop. Dis. 2015, 9, e0004217. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.J.; Alvarado, M.; Basanez, M.G.; Bolliger, I.; Bourne, R.; Boussinesq, M.; Brooker, S.J.; Brown, A.S.; Buckle, G.; Budke, C.M.; et al. The Global Burden of Disease Study 2010: Interpretation and Implications for the Neglected Tropical Diseases. PLoS Negl. Trop. Dis. 2014, 8, e2865. [Google Scholar] [CrossRef] [PubMed]
- Leenstra, T.; Acosta, L.P.; Langdon, G.C.; Manalo, D.L.; Su, L.; Olveda, R.M.; McGarvey, S.T.; Kurtis, J.D.; Friedman, J.F. Schistosomiasis japonica, anemia, and iron status in children, adolescents, and young adults in Leyte, Philippines. Am. J. Clin. Nutr. 2006, 83, 371–379. [Google Scholar] [CrossRef]
- Coutinho, H.M.; McGarvey, S.T.; Acosta, L.P.; Manalo, D.L.; Langdon, G.C.; Leenstra, T.; Kanzaria, H.K.; Solomon, J.; Wu, H.W.; Olveda, R.M.; et al. Nutritional status and serum cytokine profiles in children, adolescents, and young adults with Schistosoma japonicum-associated hepatic fibrosis, in Leyte, Philippines. J. Infect. Dis. 2005, 192, 528–536. [Google Scholar] [CrossRef]
- Soares Magalhães, R.J.; Salamat, M.S.; Leonardo, L.; Gray, D.J.; Carabin, H.; Halton, K.; McManus, D.P.; Williams, G.M.; Rivera, P.; Saniel, O.; et al. Geographical distribution of human Schistosoma japonicum infection in The Philippines: Tools to support disease control and further elimination. Int. J. Parasitol. 2014, 44, 977–984. [Google Scholar] [CrossRef]
- Herbreteau, V.; Salem, G.; Souris, M.; Hugot, J.-P.; Gonzalez, J.-P. Thirty years of use and improvement of remote sensing, applied to epidemiology: From early promises to lasting frustration. Health Place 2007, 13, 400–403. [Google Scholar] [CrossRef]
- Hay, S.I.; Packer, M.; Rogers, D. Review article The impact of remote sensing on the study and control of invertebrate intermediate hosts and vectors for disease. Int. J. Remote Sens. 1997, 18, 2899–2930. [Google Scholar] [CrossRef]
- Kalluri, S.; Gilruth, P.; Rogers, D.; Szczur, M. Surveillance of arthropod vector-borne infectious diseases using remote sensing techniques: A review. PLoS Pathog. 2007, 3, e116. [Google Scholar] [CrossRef] [PubMed]
- Hamm, N.A.S.; Soares Magalhães, R.J.; Clements, A.C.A. Earth Observation, Spatial Data Quality, and Neglected Tropical Diseases. PLoS Negl. Trop. Dis. 2015, 9, e0004164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Manjourides, J.; Cohen, T.; Hu, Y.; Jiang, Q.W. Spatial measurement errors in the field of spatial epidemiology. Int. J. Health Geogr. 2016, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- Araujo Navas, A.L.; Hamm, N.A.S.; Soares Magalhães, R.J.; Stein, A. Mapping Soil Transmitted Helminths and Schistosomiasis under Uncertainty: A Systematic Review and Critical Appraisal of Evidence. PLoS Negl. Trop. Dis. 2016, 10, e0005208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Goodchild, M.F. Uncertainty in Geographical Information; Taylor & Francis Group: London, UK, 2002; ISBN 0-415-27723-X. [Google Scholar]
- Richardson, S.; Monfort, C. Ecological Correlation Studies. In Spatial Epidemiology: Methods and Applications; Elliot, P.W., Wakefield, J.C., Best, N.G., Briggs, D.J., Eds.; Oxford University Press: Oxford, UK, 2000; pp. 205–220. ISBN 0192629417. [Google Scholar]
- Wakefield, J.; Shaddick, G. Health-exposure modeling and the ecological fallacy. Biostatistics 2006, 7, 438–455. [Google Scholar] [CrossRef] [PubMed]
- King, G. A Solution to the Ecological Inference Problem: Reconstructing Individual Behavior from Aggregate Data; Princeton University Press: Princeton, NJ, USA, 2013; ISBN 1400849209. [Google Scholar]
- Wakefield, J.; Lyons, H. Spatial Aggregation and the Ecological Fallacy. In Handbook of Modern Statisticcal Methods; Hall/CRC, C., Ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 541–558. ISBN 9781420072877. [Google Scholar]
- Gelfand, A.E.; Diggle, P.; Guttorp, P.; Fuentes, M. Handbook of Spatial Statistics; Taylor & Francis Group: Boca Raton, FL, USA, 2010; p. 619. ISBN 9781420072877. [Google Scholar]
- Prentice, R.L.; Sheppard, L. Aggregate data studies of disease risk factors. Biometrika 1995, 82, 113–125. [Google Scholar] [CrossRef]
- Richardson, S.; Stucker, I.; Hemon, D. Comparison of relative risks obtained in ecological and individual studies: Some methodological considerations. Int. J. Epidemiol. 1987, 16, 111–120. [Google Scholar] [CrossRef]
- Wang, F.F.; Wang, J.; Gelfand, A.; Li, F. Accommodating the ecological fallacy in disease mapping in the absence of individual exposures. Stat. Med. 2017, 36, 4930–4942. [Google Scholar] [CrossRef] [Green Version]
- Raso, G.; Matthys, B.; N’goran, E.; Tanner, M.; Vounatsou, P.; Utzinger, J. Spatial risk prediction and mapping of Schistosoma mansoni infections among schoolchildren living in western Côte d’Ivoire. Parasitology 2005, 131, 97–108. [Google Scholar] [CrossRef]
- Chammartin, F.; Houngbedji, C.A.; Huerlimann, E.; Yapi, R.B.; Silue, K.D.; Soro, G.; Kouame, F.N.; N’Goran, E.K.; Utzinger, J.; Raso, G.; et al. Bayesian Risk Mapping and Model-Based Estimation of Schistosoma haematobium-Schistosoma mansoni Co-distribution in Cote d’Ivoire. PLoS Negl. Trop. Dis. 2014, 8, e3407. [Google Scholar] [CrossRef] [PubMed]
- Woodhall, D.M.; Wiegand, R.E.; Wellman, M.; Matey, E.; Abudho, B.; Karanja, D.M.S.; Mwinzi, P.M.N.; Montgomery, S.P.; Secor, W.E. Use of Geospatial Modeling to Predict Schistosoma mansoni Prevalence in Nyanza Province, Kenya. PLoS ONE 2013, 8, e71635. [Google Scholar] [CrossRef]
- Sturrock, H.J.W.; Pullan, R.L.; Kihara, J.H.; Mwandawiro, C.; Brooker, S.J. The Use of Bivariate Spatial Modeling of Questionnaire and Parasitology Data to Predict the Distribution of Schistosoma haematobium in Coastal Kenya. PLoS Negl. Trop. Dis. 2013, 7, e2016. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Bergquist, R.; Lynn, H.; Gao, F.; Wang, Q.; Zhang, S.; Li, R.; Sun, L.; Xia, C.; Xiong, C.; et al. Sandwich mapping of schistosomiasis risk in Anhui Province, China. Geospat. Health 2015, 10, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Souza Guimaraes, R.J.d.P.; Freitas, C.C.; Dutra, L.V.; Carvalho Scholte, R.G.; Martins-Bede, F.T.; Fonseca, F.R.; Amaral, R.S.; Drummonds, S.C.; Felgueiras, C.A.; Oliveira, G.C.; et al. A geoprocessing approach for studying and controlling schistosomiasis in the state of Minas Gerais, Brazil. Mem. Inst. Oswaldo Cruz 2010, 105, 524–531. [Google Scholar] [CrossRef] [Green Version]
- Scholte, R.G.C.; Gosoniu, L.; Malone, J.B.; Chammartin, F.; Utzinger, J.; Vounatsou, P. Predictive risk mapping of schistosomiasis in Brazil using Bayesian geostatistical models. Acta Trop. 2014, 132, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Soares Magalhães, R.J.; Salamat, M.S.; Leonardo, L.; Gray, D.J.; Carabin, H.; Halton, K.; McManus, D.P.; Williams, G.M.; Rivera, P.; Saniel, O.; et al. Mapping the Risk of Soil-Transmitted Helminthic Infections in the Philippines. PLoS Negl. Trop. Dis. 2015, 9, e0003915. [Google Scholar] [CrossRef]
- Clements, A.C.A.; Brooker, S.; Nyandindi, U.; Fenwick, A.; Blair, L. Bayesian spatial analysis of a national urinary schistosomiasis questionnaire to assist geographic targeting of schistosomiasis control in Tanzania, East Africa. Int. J. Parasitol. 2008, 38, 401–415. [Google Scholar] [CrossRef] [Green Version]
- Leonardo, L.; Rivera, P.; Saniel, O.; Villacorte, E.; Lebanan, M.A.; Crisostomo, B.; Hernandez, L.; Baquilod, M.; Erce, E.; Martinez, R. A national baseline prevalence survey of schistosomiasis in the Philippines using stratified two-step systematic cluster sampling design. J. Trop. Med. 2012, 2012, 8. [Google Scholar] [CrossRef]
- Leonardo, L.; Rivera, P.; Saniel, O.; Solon, J.A.; Chigusa, Y.; Villacorte, E.; Chua, J.C.; Moendeg, K.; Manalo, D.; Crisostomo, B.; et al. New endemic foci of schistosomiasis infections in the Philippines. Acta Trop. 2015, 141, 354–360. [Google Scholar] [CrossRef]
- Leonardo, L.; Acosta, L.P.; Olveda, R.M.; Aligui, G.D.L. Difficulties and strategies in the control of schistosomiasis in the Philippines. Acta Trop. 2002, 82, 295–299. [Google Scholar] [CrossRef]
- Zhou, X.N.; Bergquist, R.; Leonardo, L.; Yang, G.J.; Yang, K.; Sudomo, M.; Olveda, R. Schistosomiasis Japonica: Control and Research Needs. In Important Helminth Infections in Southeast Asia: Diversity and Potential for Control and Elimination, Peart A; Zhou, X.N., Bergquist, R., Olveda, R., Utzinger, J., Eds.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2010; Volume 72, pp. 145–178. ISBN 978-0-12-381513-2. [Google Scholar]
- Leonardo, L.R.; Rivera, P.; Saniel, O.; Villacorte, E.; Crisostomo, B.; Hernandez, L.; Baquilod, M.; Erce, E.; Martinez, R.; Velayudhan, R. Prevalence survey of schistosomiasis in Mindanao and the Visayas, The Philippines. Parasitol. Int. 2008, 57, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.L.N.; Cerqueira, E.J.L.; Soares, N.M. Comparison of the thick smear and Kato-Katz techniques for diagnosis of intestinal helminth infections. Rev. Soc. Bras. Med. Trop. 2005, 38, 196–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ESRI. ArcGIS Desktop, 10; Environmental Systems Research Institute: Redlands, CA, USA, 2011. [Google Scholar]
- Zhou, Y.B.; Liang, S.; Jiang, Q.W. Factors impacting on progress towards elimination of transmission of schistosomiasis japonica in China. Parasit. Vectors 2012, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Walz, Y.; Wegmann, M.; Dech, S.; Raso, G.; Utzinger, J. Risk profiling of schistosomiasis using remote sensing: Approaches, challenges and outlook. Parasit. Vectors 2015, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Xu, H. Modification of normalised difference water index (NDWI) to enhance open water features in remotely sensed imagery. Int. J. Remote Sens. 2006, 27, 3025–3033. [Google Scholar] [CrossRef]
- Prah, S.; James, C. The influence of physical factors on the survival and infectivity of miracidia of Schistosoma mansoni and S. haematobium I. Effect of temperature and ultra-violet light. J. Helminthol. 1977, 51, 73–85. [Google Scholar] [CrossRef]
- Woolhouse, M.; Chandiwana, S. Population dynamics model for Bulinus globosus, intermediate host for Schistosoma haematobium, in river habitats. Acta Trop. 1990, 47, 151–160. [Google Scholar] [CrossRef]
- Pietrock, M.; Marcogliese, D.J. Free-living endohelminth stages: At the mercy of environmental conditions. Trends Parasitol. 2003, 19, 293–299. [Google Scholar] [CrossRef]
- Geological Survey, U.S. Global Data Explorer. Available online: https://gdex.cr.usgs.gov/gdex/ (accessed on 15 August 2017).
- Pesigan, T.P.; Hairston, N.G.; Jauregui, J.J.; Garcia, E.G.; Santos, A.T.; Santos, B.C.; Besa, A.A. Studies on Schistosoma japonicum infection in the Philippines 2. The molluscan host. Bull. World Health Organ. 1958, 18, 481–578. [Google Scholar]
- Stensgaard, A.S.; Jorgensen, A.; Kabatereine, N.B.; Rahbek, C.; Kristensen, T.K. Modeling freshwater snail habitat suitability and areas of potential snail-borne disease transmission in Uganda. Geospat. Health 2006, 1, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Stensgaard, A.S.; Utzinger, J.; Vounatsou, P.; Hurlimann, E.; Schur, N.; Saarnak, C.F.L.; Simoonga, C.; Mubita, P.; Kabatereine, N.B.; Tchuente, L.A.T.; et al. Large-scale determinants of intestinal schistosomiasis and intermediate host snail distribution across Africa: Does climate matter? Acta Trop. 2013, 128, 378–390. [Google Scholar] [CrossRef] [PubMed]
- National Mapping and Resource Information Authority. Available online: http://www.namria.gov.ph/ (accessed on 15 February 2018).
- Gelman, A. Prior distributions for variance parameters in hierarchical models (Comment on an Article by Browne and Draper). Bayesian Anal. 2006, 1, 515–533. [Google Scholar] [CrossRef]
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Rubin, D.B. Bayesian Data Analysis; Chapman and Hall/CRC: Boca Raton, FL, USA, 1995; ISBN 1135439419. [Google Scholar]
- Diggle, P.J.; Tawn, J.; Moyeed, R. Model-based geostatistics. J. R. Stat. Soc. Ser. C Appl. Stat. 2002, 47, 299–350. [Google Scholar] [CrossRef]
- Thomas, A.; Best, N.; Lunn, D.; Arnold, R.; Spiegelhalter, D. GeoBugs User Manual; MRC Biostatistics Unit: Cambridge, UK, 2004. [Google Scholar]
- Lunn, D.; Jackson, C.; Best, N.; Thomas, A.; Spiegelhalter, D. The BUGS Book: A Practical Introduction to Bayesian Analysis; CRC Press: Boca Raton, FL, USA, 2012; ISBN 1584888490. [Google Scholar]
- Brooks, S.P.; Gelman, A. General methods for monitoring convergence of iterative simulations. J. Comput. Graph. Stat. 1998, 7, 434–455. [Google Scholar] [CrossRef]
- Gelman, A.; Rubin, D.B. Inference from iterative simulation using multiple sequences. Stat. Sci. 1992, 7, 457–472. [Google Scholar] [CrossRef]
- Brooker, S.; Hay, S.I.; Bundy, D.A.P. Tools from ecology: Useful for evaluating infection risk models? Trends Parasitol. 2002, 18, 70–74. [Google Scholar] [CrossRef]
- Hijmans, R.; Rojas, E.; Cruz, M.; O’Brien, R.; Barrantes, I. DIVA-GIS free, simple and effective. Available online: http://www.diva-gis.org/Data (accessed on 8 April 2018).
- Project, O.S.M. Planet OSM; Open Street Map Project: Cambridge, UK, 2017. [Google Scholar]
- OCHA Humanitarian Data Exchange v1.25.3. Available online: https://data.humdata.org/search?groups=phl&q=&ext_page_size=25 (accessed on 10 April 2018).
- Spiegelhalter, D.; Thomas, A.; Best, N.; Lunn, D. OpenBUGS User Manual; Version 3.0.2.; MRC Biostatistics Unit: Cambridge, UK, 2007. [Google Scholar]
- Spiegelhalter, D.; Thomas, A.; Best, N.; Lunn, D. WinBUGS User Manual; MRC Biostatistics Unit: Cambridge, UK, 2003. [Google Scholar]
- Lunn, D.; Spiegelhalter, D.; Thomas, A.; Best, N. OpenBUGS license. Available online: http://www.openbugs.net/w/Downloads (accessed on 25 June 2018).
- Sturtz, S.; Ligges, U.; Gelman, A. R2OpenBUGS: A Package for Running OpenBUGS from R.; Journal of Statistical Software: Innsbruck, Austria, 2010. [Google Scholar]
- Team, R.D.C. R: A Language and Environment for Statistical Computing; The R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Brooker, S.; Hay, S.; Issae, W.; Hall, A.; Kihamia, C.; Lawambo, N.; Wint, W.; Rogers, D.; Bundy, D. Predicting the distribution of urinary schistosomiasis in Tanzania using satellite sensor data. Trop. Med. Int. Health 2001, 6, 998–1007. [Google Scholar] [CrossRef] [Green Version]
- Kristensen, T.; Malone, J.; McCarroll, J. Use of satellite remote sensing and geographic information systems to model the distribution and abundance of snail intermediate hosts in Africa: A preliminary model for Biomphalaria pfeifferi in Ethiopia. Acta Trop. 2001, 79, 73–78. [Google Scholar] [CrossRef]
- Brooker, S.; Hay, S.I.; Tchuente, L.-A.T.; Ratard, R. Using NOAA-AVHRR data to model human helminth distributions in planning disease control in Cameroon, West Africa. Photogramm. Eng. Remote Sens. 2002, 68, 175–179. [Google Scholar]
- Malone, J.B.; Yilma, J.M.; McCarroll, J.C.; Erko, B.; Mukaratirwa, S.; Zhou, X.Y. Satellite climatology and the environmental risk of Schistosoma mansoni in Ethiopia and east Africa. Acta Trop. 2001, 79, 59–72. [Google Scholar] [CrossRef]
- Rice Science for a Better World. Available online: http://irri.org/our-work/research/policy-and-markets/mapping-rice-in-the-philippines-where (accessed on 18 October 2018).
- Stensgaard, A.; Jorgensen, A.; Kabatareine, N.; Malone, J.; Kristensen, T. Modeling the distribution of Schistosoma mansoni and host snails in Uganda using satellite sensor data and Geographical Information Systems. Parassitologia 2005, 47, 115–125. [Google Scholar] [PubMed]
- Burns, C.J.; Wright, M.; Pierson, J.B.; Bateson, T.F.; Burstyn, I.; Goldstein, D.A.; Klaunig, J.E.; Luben, T.J.; Mihlan, G.; Ritter, L.; et al. Evaluating Uncertainty to Strengthen Epidemiologic Data for Use in Human Health Risk Assessments. Environ. Health Perspect. 2014, 122, 1160–1165. [Google Scholar] [CrossRef] [Green Version]
- Schur, N.; Hurlimann, E.; Garba, A.; Traore, M.S.; Ndir, O.; Ratard, R.C.; Tchuente, L.A.T.; Kristensen, T.K.; Utzinger, J.; Vounatsou, P. Geostatistical Model-Based Estimates of Schistosomiasis Prevalence among Individuals Aged <= 20 Years in West Africa. PLoS Negl. Trop. Dis. 2011, 5, e1194. [Google Scholar] [CrossRef] [PubMed]
- Schur, N.; Hurlimann, E.; Stensgaard, A.S.; Chimfwembe, K.; Mushinge, G.; Simoonga, C.; Kabatereine, N.B.; Kristensen, T.K.; Utzinger, J.; Vounatsou, P. Spatially explicit Schistosoma infection risk in eastern Africa using Bayesian geostatistical modelling. Acta Trop. 2013, 128, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Simoonga, C.; Utzinger, J.; Brooker, S.; Vounatsou, P.; Appleton, C.C.; Stensgaard, A.S.; Olsen, A.; Kristensen, T.K. Remote sensing, geographical information system and spatial analysis for schistosomiasis epidemiology and ecology in Africa. Parasitology 2009, 136, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- White, H. A heteroskedasticity-consistent covariance matrix estimator and a direct test for heteroskedasticity. Econometrica 1980, 817–838. [Google Scholar] [CrossRef]
- Fox, J. Applied Regression Analysis, Linear Models, and Related Methods; Sage Publications, Inc.: Thousand Oaks, CA, USA, 1997; ISBN 080394540X. [Google Scholar]
- Mankiw, N.G. A Quick Refresher Course in Macroeconomics. J. Econ. Lit. 1990, 28, 1645–1660. [Google Scholar]

| Environmental Variable | Spatial Resolution | Temporal Resolution | Data Type | Original Coordinate System | Data Source |
|---|---|---|---|---|---|
| Elevation | 30 m | NA | Raster | EPSG:4326 | Aster GDEM V2 from USGS |
| NDVI | 250 m | 2008 | Raster | EPSG:4326 | MOD13Q1 |
| NDWI | 500 m | 2008 | Raster | EPSG:32651 | Landsat 7, one-year composite |
| LST | 1 km | 2008 | Raster | EPSG:4326 | MOD11A2 |
| NDWB | 250 m | 2010 | Raster | EPSG:32651 | Derived from closest facility network using roads, urban areas, river network, and water bodies |
| Estimated Parameters | Posterior Mean (95% Crl) | Standard Deviation | Credible Intervals Width (Uncertainty) | |||
|---|---|---|---|---|---|---|
| Convolution Model | Ecological Model | Convolution Model | Ecological Model | Convolution Model | Ecological Model | |
| Intercept | −5.79 (−6.11,−5.5) | −5.67 (−5.93,−5.41) | 0.16 | 0.13 | 0.62 | 0.53 |
| NDWI | −0.74 (−0.96,−0.55) | −1.02 (−1.24,−0.80) | 0.11 | 0.11 | 0.44 | 0.44 |
| LSTD | −0.63 (−0.92,−0.38) | −0.79 (−1.08,−0.49) | 0.14 | 0.15 | 0.56 | 0.59 |
| LSTN | −0.84 (−1.13,−0.55) | −0.65 (−1.05,−0.24) | 0.15 | 0.21 | 0.59 | 0.82 |
| Elevation | −1.05 (−1.4,−0.71) | −1.13 (−1.53,−0.70) | 0.18 | 0.22 | 0.71 | 0.84 |
| NDWB | −0.28 (−0.51,−0.05) | −0.24 (−0.43,−0.05) | 0.13 | 0.09 | 0.48 | 0.38 |
| ϕ | 4 × 10−5 (−0.004,0.004) | 2 × 10−5 (−0.0004,0.0004) | 2.00 × 10−4 | 1.00 × 10−5 | 6.80 × 10−5 | 3.50 × 10−5 |
| Variance of spatial random effect | 2.58 (1.7,3.6) | 2.6 (1.8,3.61) | 0.48 | 0.47 | 1.9 | 1.82 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araujo Navas, A.L.; Osei, F.; Leonardo, L.R.; Soares Magalhães, R.J.; Stein, A. Modeling Schistosoma japonicum Infection under Pure Specification Bias: Impact of Environmental Drivers of Infection. Int. J. Environ. Res. Public Health 2019, 16, 176. https://doi.org/10.3390/ijerph16020176
Araujo Navas AL, Osei F, Leonardo LR, Soares Magalhães RJ, Stein A. Modeling Schistosoma japonicum Infection under Pure Specification Bias: Impact of Environmental Drivers of Infection. International Journal of Environmental Research and Public Health. 2019; 16(2):176. https://doi.org/10.3390/ijerph16020176
Chicago/Turabian StyleAraujo Navas, Andrea L., Frank Osei, Lydia R. Leonardo, Ricardo J. Soares Magalhães, and Alfred Stein. 2019. "Modeling Schistosoma japonicum Infection under Pure Specification Bias: Impact of Environmental Drivers of Infection" International Journal of Environmental Research and Public Health 16, no. 2: 176. https://doi.org/10.3390/ijerph16020176
APA StyleAraujo Navas, A. L., Osei, F., Leonardo, L. R., Soares Magalhães, R. J., & Stein, A. (2019). Modeling Schistosoma japonicum Infection under Pure Specification Bias: Impact of Environmental Drivers of Infection. International Journal of Environmental Research and Public Health, 16(2), 176. https://doi.org/10.3390/ijerph16020176








