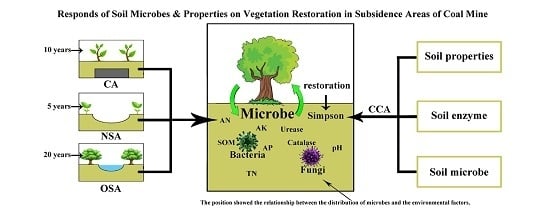
Response of Soil Microbes to Vegetation Restoration in Coal Mining Subsidence Areas at Huaibei Coal Mine, China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Soil Sample Collection
2.3. Measurement of Soil Parameters
2.4. Data Analysis
3. Results
3.1. Composition of Soil Microorganism Community
3.1.1. Dominant Bacterial Genera in Soil Microorganism Community
3.1.2. Soil Microbial Diversity
3.2. Analysis of Soil Enzyme Activity
3.3. CCA Ordination of Soil Microorganisms with Environmental Factors in the Soils
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hu, Z.Q.; Wang, X.J.; He, A.M. Distribution characteristic and development rules of ground fissures due to coal mining in windy and sandy region. J. China Coal Soc. 2014, 39, 11–18. [Google Scholar] [CrossRef]
- Zhou, D.W.; Wu, K.; Cheng, G.L.; Li, L. Mechanism of mining subsidence in coal mining area with thick alluvium soil in China. Arab. J. Geosci. 2015, 8, 1855–1867. [Google Scholar] [CrossRef]
- Huang, J.; Tian, C.Y.; Xing, L.F.; Bian, Z.F.; Miao, X.X. Green and Sustainable Mining: Underground Coal Mine Fully Mechanized Solid Dense Stowing-Mining Method. Sustainability 2017, 9, 1418. [Google Scholar] [CrossRef]
- Bi, Y.L. Research advance of application of arbuscular mycorrhizal fungi to ecological remediation in subsided land of coal mining areas. Mycosystema 2017, 36, 800–806. [Google Scholar] [CrossRef]
- Cheng, W.; Bian, Z.F.; Dong, J.H.; Lei, S.G. Soil properties in reclaimed farmland by filling subsidence basin due to underground coal mining with mineral wastes in China. Trans. Nonferrous Met. Soc. China 2014, 24, 2627–2635. [Google Scholar] [CrossRef]
- Gilland, K.E.; McCarthy, B.C. Microtopography influences early successional plant communities on experimental coal surface mine land reclamation. Restor. Ecol. 2014, 22, 232–239. [Google Scholar] [CrossRef]
- Wright, I.; McCarthy, B.; Belmer, N.; Price, P. Subsidence from an underground coal mine and mine wastewater discharge causing water pollution and degradation of aquatic ecosystems. Water Air Soil Pollut. 2015, 226, 1–14. [Google Scholar] [CrossRef]
- Wang, Q.N.; Chen, X.S.; Zhang, Z.G. Analysis on the status guo, problems and reasons of the reclamation for coal mine subsidence areas in China. Energy Environ. Prot. 2008, 22, 49–53. [Google Scholar] [CrossRef]
- Zang, Y.T.; Wang, J.; Ding, G.D.; Gao, Y.; He, X.; Yan, L.; He, Z.; Na, Q.; Gong, P.; Ren, Y. Variation of physico-chemical properties of aeolian sandy soil at coal mining subsidence and its evaluation. Acta Pedol. Sin. 2010, 47, 262–269. [Google Scholar] [CrossRef]
- Hu, Z.Q.; Chen, C. Impact of underground coal mining on land ecology and its restoration in windy and sandy region. J. Min. Sci. Technol. 2016, 1, 120–130. [Google Scholar] [CrossRef]
- Walker, L.; Walker, J.; Hobbs, R.J. Linking Restoration and Ecological Succession; Springer: New York, NY, USA, 2007. [Google Scholar]
- An, S.S.; Huang, Y.M.; Zheng, F.L. Evaluation of soil microbial indices along a revegetation chronosequence in grassland soils on the Loess Plateau, Northwest China. Appl. Soil Ecol. 2009, 41, 286–292. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.; Xue, S.; Wang, G. Changes in rhizospheric microbial community structure and function during the natural recovery of abandoned cropland on the Loess Plateau, China. Ecol. Eng. 2015, 75, 161–171. [Google Scholar] [CrossRef]
- Putten, W.H.; Bardgett, R.D.; Bever, J.D.; Bezemer, T.M.; Casper, B.B.; Fukami, T.; Kardol, P.; Klironomos, J.; Kulmatiski, A.; Schweitzer, J.A.; et al. Plant-soil feedbacks: The past, the present and future challenges. J. Ecol. 2013, 101, 265–276. [Google Scholar] [CrossRef]
- Waldrop, M.P.; Firestone, M.K. Response of Microbial Community Composition and Function to Soil Climate Change. Microb. Ecol. 2006, 52, 716–724. [Google Scholar] [CrossRef]
- Yao, H.; He, Z.; Wilson, M.J. Microbial biomass and community structure in a sequence of soils with increasing fertility and changing land use. Microb. Ecol. 2000, 40, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Liu, F.; Zhou, X.M. Effects of different reclaimed scenarious on soil microbe and enzyme activities in mining area. Environ. Sci. 2015, 36, 1836–1841. [Google Scholar] [CrossRef]
- Dimitriu, P.A.; Prescott, C.E.; Quideau, S.A.; Grayston, S.J. Impact of reclamation of surface-mined boreal forest soils on microbial community composition and function. Soil Biol. Biochem. 2010, 42, 2289–2297. [Google Scholar] [CrossRef]
- Dangi, S.R.; Stahl, P.D.; Wick, A.F.; Ingram, L.J.; Buyer, J.S. Soil microbial community recovery in reclaimed soilson a surface coal mine site. Soil Sci. Soc. Am. J. 2012, 76, 915–924. [Google Scholar] [CrossRef]
- Kardol, P.; Bezemer, T.M.; van der Putten, W.H. Temporal variation in plant-soil feedback controls succession. Ecol. Lett. 2006, 9, 1080–1088. [Google Scholar] [CrossRef]
- Kuramae, E.; Gamper, H.; Van, V.J.; Kowalchuk, G. Soil and plant factors driving the community of soil-borne microorganisms across chronosequences of secondary succession of chalk grasslands with a neutral pH. FEMS Microbiol. Ecol. 2011, 77, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Li, J.J.; Zheng, Y.M.; Yan, J.X.; Li, H.J.; He, J.Z. Succession of plant and soil microbial communities with restoration of abandoned land in the Loess Plateau, China. J. Soils Sediments 2013, 13, 760–769. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.K.; Huang, X.H.; Duan, C.H.; He, S.Z.; Zhang, G.S.; Liu, C.E.; Fu, D.G.; Shao, H.B. Impacts of Ageratina adenophora invasion on soil physical–chemical properties of Eucalyptus plantation and implications for constructing agro-forest ecosystem. Ecol. Eng. 2014, 64, 130–135. [Google Scholar] [CrossRef]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; Gonzálezet, A.; Caporaso, J.G.; Knight, R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinform. 2011. [Google Scholar] [CrossRef]
- Liu, R.; Xiao, N.; Wei, S.H.; Zhao, L.X.; An, J. Rhizosphere effects of PAH-contaminated soil phytoremediation using a special plant named Fire Phoenix. Sci. Total Environ. 2014, 473–474, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Kandeler, E.; Gerber, H. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fertil. Soils 1988, 6, 68–72. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, J.F.; Xu, H.S.; Chen, G.C.; Wang, L.P. Variation of soil microbial community composition and enzyme activities with different salinities on Yuyao coast, Zhejiang, China. Chin. J. Appl. Ecol. 2016, 27, 3361–3370. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 11 March 2019).
- Pester, M.; Maixner, F.; Berry, D.; Rattei, T.; Koch, H.; Lücker, S.; Nowka, B.; Richter, A.; Spieck, E.; Lebedeva, E.; et al. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 2013, 16. [Google Scholar] [CrossRef]
- Shun, H.; Luyang, Z.; Xuesong, L.; Xiong, X.; Wen, S.; Wang, B.; Chen, W.; Huang, Q. Shifts in, Nitrobacter- and, Nitrospira-like nitrite-oxidizing bacterial communities under long-term fertilization practices. Soil Biol. Biochem. 2018, 124, 118–125. [Google Scholar] [CrossRef]
- Tobin, T.; Shade, A.; Marshall, L.J.; Torres, K.; Beblo, C.; Janzen, C.; Lenig, J.; Martinez, A.; Ressler, D. Nitrogen changes and domain bacteria ribotype diversity in soils overlying the Centralia, Pennsylvania underground coal mine fire. Soil Sci. 2005, 170, 191–201. [Google Scholar] [CrossRef]
- Aditiawati, P.; Akhmaloka Astuti, D.I.; Sugilubin Pikoli, M.R. Biodesulfurization of subbituminous coal by mixed culture bacteria isolated from coal mine soil of South Sumatera. Biotechnology 2013, 12, 46–53. [Google Scholar] [CrossRef]
- Beneduzi, A.; Peres, D.; Da, C.P.; Bodanese Zanettini, M.H.; Passaglia, L.M. Genetic and phenotypic diversity of plant-growth-promoting bacilli isolated from wheat fields in southern Brazil. Res. Microbiol. 2008, 159, 244–250. [Google Scholar] [CrossRef]
- Chen, L.X.; Li, J.T.; Chen, Y.T.; Huang, N.L.; Hua, Z.S.; Hu, M.; Shu, W.S. Shifts in microbial community composition and function in the acidification of a lead/zinc mine tailings. Environ. Microbiol. 2013, 15, 2431–2444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Chen, L.; Wen, H. Changes in the composition and diversity of bacterial communities 13 years after soil reclamation of abandoned mine land in eastern China. Ecol. Res. 2015, 30, 357–366. [Google Scholar] [CrossRef]
- Rich, J.J.; Myrold, D.D. Community composition and activities of denitrifying bacteria from adjacent agricultural soil, riparian soil, and creek sediment in oregon, usa. Soil Biol. Biochem. 2004, 36, 1431–1441. [Google Scholar] [CrossRef]
- Keil, D.; Meyer, A.; Berner, D.; Poll, C.; Schützenmeister, A.; Piepho, H.P.; Vlasenko, A.; Philippot, L.; Schloter, M.; Kandeler, E.; Marhan, S. Influence of land-use intensity on the spatial distribution of n-cycling microorganisms in grassland soils. FEMS Microbiol. Ecol. 2011, 77, 95–106. [Google Scholar] [CrossRef]
- Wu, X.; Monchy, S.; Taghavi, S.; Zhu, W.; Ramos, J.; Lelie, D.V.D. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol. Rev. 2011, 35, 299–323. [Google Scholar] [CrossRef]
- Ma, Q.; Qu, Y.Y.; Zhang, X.W.; Shen, W.L.; Liu, Z.Y.; Wang, J.W.; Zhang, Z.J.; Zhou, J.T. Identification of the microbial community composition and structure of coal-mine wastewater treatment plants. Microbiol. Res. 2015, 175, 1–5. [Google Scholar] [CrossRef]
- Ye, R.W.; Thomas, S.M. Microbial nitrogen cycles: Physiology, genomics and applications. Curr. Opin. Microbiol. 2001, 4, 307–312. [Google Scholar] [CrossRef]
- Lee, E.H.; Eo, J.K.; Lee, C.S.; Eom, A.H. Effect of soil Ameliorators on ectomycorrhizal fungal communities that colonize seedlings of Pinus densiflora in abandoned coal mine spoils. Mycobiology 2012, 40, 168–172. [Google Scholar] [CrossRef]
- Kang, H.Z.; Gao, H.H.; Yu, W.J.; Yi, Y.; Wang, Y.; Ning, M.L. Changes in soil microbial community structure and function after afforestation depend on species and age: Case study in a subtropical alluvial island. Sci. Total Environ. 2018, 625, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhou, P.; Mao, L.; Zhi, Y.; Zhang, C.; Shi, W. Effects of plant species coexistence on soil enzyme activities and soil microbial community structure under Cd and Pb combined pollution. Acta Sci. Circumstantiae 2010, 22, 1040–1048. [Google Scholar] [CrossRef]
- Wang, R.; Ma, S.C.; Zhang, H.B.; Xu, C.Y.; Guo, Z.Z. Effects of Surface Cracks Caused by High Intensity Coal Mining on Soil Microbial Characteristics and Plant Communities in Arid Regions. Res. Environ. Sci. 2016, 29, 1249–1255. [Google Scholar] [CrossRef]
- Zhang, L.J.; Wang, H.L.; Hu, B.; Li, D.Y. Analysis of the soil nutrition and enzyme activity and their correlations in the coal mining subsidence area of JiaoZuo city, Henan province―In the case of the subsidence area of Han Wang zhuang mine. Environ. Sci. Manag. 2007, 32, 126–129. [Google Scholar] [CrossRef]
- Zaman, M.; Cameron, K.C.; Di, H.J.; Inubushi, K. Changes in mineral N, microbial biomass and enzyme activities in different soil depths after surface applications of dairy shed effluent and ammonium fertilizer. Nutr. Cycl. Agroecosyst. 2002, 63, 275–290. [Google Scholar] [CrossRef]
- Hopfensperger, K.N.; Burgin, A.J.; Schoepfer, V.A.; Helton, A.M. Impacts of Saltwater Incursion on Plant Communities, Anaerobic Microbial Metabolism, and Resulting Relationships in a Restored Freshwater Wetland. Ecosystems 2014, 17, 792–807. [Google Scholar] [CrossRef]
- Floch, C.; Capowiez, Y.; Criquet, S. Enzyme activities in apple orchard agroecosystems: How are they affected by management strategy and soil properties. Soil Biol. Biochem. 2009, 41, 61–68. [Google Scholar] [CrossRef]
- Xie, X.; Pu, L.; Wang, Q.; Zhu, M.; Xu, Y.; Zhang, M. Response of soil physicochemical properties and enzyme activities to long-term reclamation of coastal saline soil, eastern China. Sci. Total Environ. 2017, 607–608, 1419. [Google Scholar] [CrossRef] [PubMed]






| Sample plots | Altitude (m) | Vegetation Restoration Time (years) | Canopy Density | Main Tree Species | Tree Height (m) | DBH (cm) |
|---|---|---|---|---|---|---|
| CA① | 36 | 10 | 0.5 | Populus sp.; Melia azedarach; Paulownia; Salix babylonica; Platycladus orientalis; Robinia pseudoacacia; Broussonetia papyrifera; Amygdalus persica; Diospyros kaki | 3–5 | 4–6 |
| NSA② | 32 | 5 | 0.7 | Populus sp.; Salix babylonica; Robinia pseudoacacia; Ulmus pumila; Ailanthus altissima; Broussonetia papyrifera; Amygdalus persica | 5–8 | 6–10 |
| OSA③ | 30 | 20 | 0.8 | Metasequoia glyptostroboides; Sabina chinensis; Koelreuteria paniculata; Ligustrum lucidum; Camptotheca acuminate; Ginkgo biloba; Broussonetia papyrifera; Trachycarpus fortunei | 8–15 | 16–24 |
| Sample Plots | Depth (cm) | pH | SOM (g·kg−1) | TN (g·kg−1) | AN (mg·kg−1) | AP (mg·kg−1) | AK (mg·kg−1) |
|---|---|---|---|---|---|---|---|
| CA1 | 0–20 | 8.18 ± 0.11aA | 11.08 ± 5.55aA | 0.42 ± 0.01aC | 62.71 ± 5.22aA | 5.76 ± 0.23aB | 186.58 ± 1.65aB |
| CA2 | 20–40 | 8.22 ± 0.04aB | 9.23 ± 3.31aA | 0.22 ± 0.01bC | 17.10 ± 0.00cB | 0.34 ± 0.30bC | 50.61 ± 0.38cC |
| CA3 | 40–60 | 8.19 ± 0.02aC | 7.02 ± 1.47aA | 0.20 ± 0.00cC | 26.22 ± 3.95bB | 0.40 ± 0.20bB | 54.21 ± 0.77bC |
| NSA1 | 0–20 | 8.30 ± 0.04abA | 15.33 ± 2.42aA | 0.62 ± 0.01aB | 71.83 ± 12.33aA | 8.18 ± 0.30bA | 192.55 ± 1.30aA |
| NSA2 | 20–40 | 8.25 ± 0.02bB | 9.78 ± 3.37bA | 0.45 ± 0.01cA | 33.06 ± 3.95bA | 4.45 ± 0.12cA | 127.11 ± 0.98cA |
| NSA3 | 40–60 | 8.35 ± 0.07aB | 7.19 ± 0.84bA | 0.51 ± 0.01bA | 30.78 ± 0.00bB | 13.80 ± 0.12aA | 150.86 ± 0.21bA |
| OSA1 | 0–20 | 8.30 ± 0.05cA | 9.60 ± 3.13aA | 0.87 ± 0.01aA | 68.41 ± 3.42aA | 1.97 ± 0.20aC | 119.83 ± 1.03aC |
| OSA2 | 20–40 | 8.38 ± 0.02bA | 6.65 ± 1.76aA | 0.33 ± 0.01bB | 34.21 ± 3.43cA | 0.93 ± 0.30bB | 68.52 ± 0.08bB |
| OSA3 | 40–60 | 8.48 ± 0.01aA | 8.31 ± 2.22aA | 0.27 ± 0.00cB | 41.19 ± 3.95bA | 0.47 ± 0.30bB | 63.79 ± 0.44cB |
| Sample Plots | Chao1 | Shannon | Ace | Simpson | ||||
|---|---|---|---|---|---|---|---|---|
| Bacteria | Fungi | Bacteria | Fungi | Bacteria | Fungi | Bacteria | Fungi | |
| CA1 | 11,479.21 | 810.99 | 11.17 | 6.79 | 12,373.23 | 807.25 | 0.9983 | 0.9646 |
| CA2 | 12,022.88 | 546.42 | 11.23 | 6.35 | 12,858.34 | 563.21 | 0.9986 | 0.9666 |
| CA3 | 11,893.88 | 460.43 | 11.08 | 5.60 | 12,664.56 | 461.89 | 0.9981 | 0.9081 |
| NSA1 | 13,652.12 | 1006.68 | 10.36 | 6.61 | 15,137.37 | 1006.85 | 0.9938 | 0.9548 |
| NSA2 | 15,436.82 | 538.33 | 10.76 | 4.48 | 16,585.60 | 538.22 | 0.9967 | 0.7751 |
| NSA3 | 12,449.85 | 508.02 | 10.40 | 5.04 | 13,351.64 | 516.77 | 0.9949 | 0.8117 |
| OSA1 | 13,877.33 | 909.98 | 11.23 | 6.76 | 15,230.25 | 906.49 | 0.9986 | 0.9713 |
| OSA2 | 11,348.95 | 415.34 | 10.53 | 5.53 | 12,326.90 | 415.62 | 0.9967 | 0.9247 |
| OSA3 | 13,806.82 | 553.11 | 11.04 | 6.76 | 14,967.82 | 562.23 | 0.9981 | 0.9735 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Sun, H.; Zhang, D.; Zhang, J.; Cai, Z.; Qin, G.; Song, Y. Response of Soil Microbes to Vegetation Restoration in Coal Mining Subsidence Areas at Huaibei Coal Mine, China. Int. J. Environ. Res. Public Health 2019, 16, 1757. https://doi.org/10.3390/ijerph16101757
Sun S, Sun H, Zhang D, Zhang J, Cai Z, Qin G, Song Y. Response of Soil Microbes to Vegetation Restoration in Coal Mining Subsidence Areas at Huaibei Coal Mine, China. International Journal of Environmental Research and Public Health. 2019; 16(10):1757. https://doi.org/10.3390/ijerph16101757
Chicago/Turabian StyleSun, Shiyong, Hui Sun, Deshun Zhang, Jianfeng Zhang, Zeyu Cai, Guanghua Qin, and Yumin Song. 2019. "Response of Soil Microbes to Vegetation Restoration in Coal Mining Subsidence Areas at Huaibei Coal Mine, China" International Journal of Environmental Research and Public Health 16, no. 10: 1757. https://doi.org/10.3390/ijerph16101757