Gcd Gene Diversity of Quinoprotein Glucose Dehydrogenase in the Sediment of Sancha Lake and Its Response to the Environment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description and Sample Collection
2.2. Characterization of Sediment Physicochemical Properties
2.3. DNA Extraction, PCR, and Illumina MiSeq Sequencing
2.4. The Diversity and Composition of gcd-Harboring Bacterial Communities Were Assessed Using High-Throughput Sequencing Technique
2.5. Statistical Analysis
3. Results and Discussion
3.1. Physicochemical Properties of the Sediments and Overlying Water
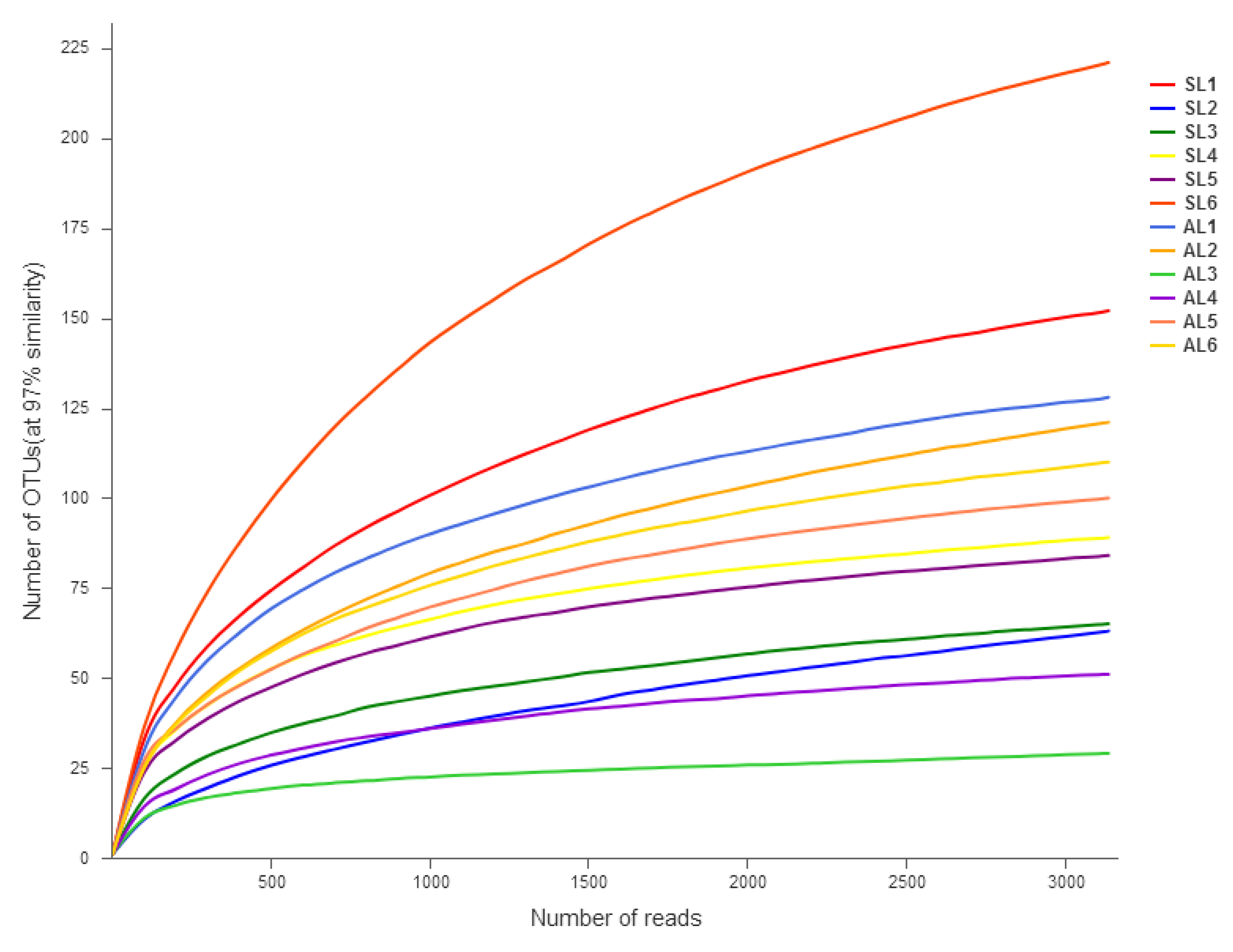
3.2. OTU Classification and Determination and Alpha Diversity of the gcd Gene in Sediments
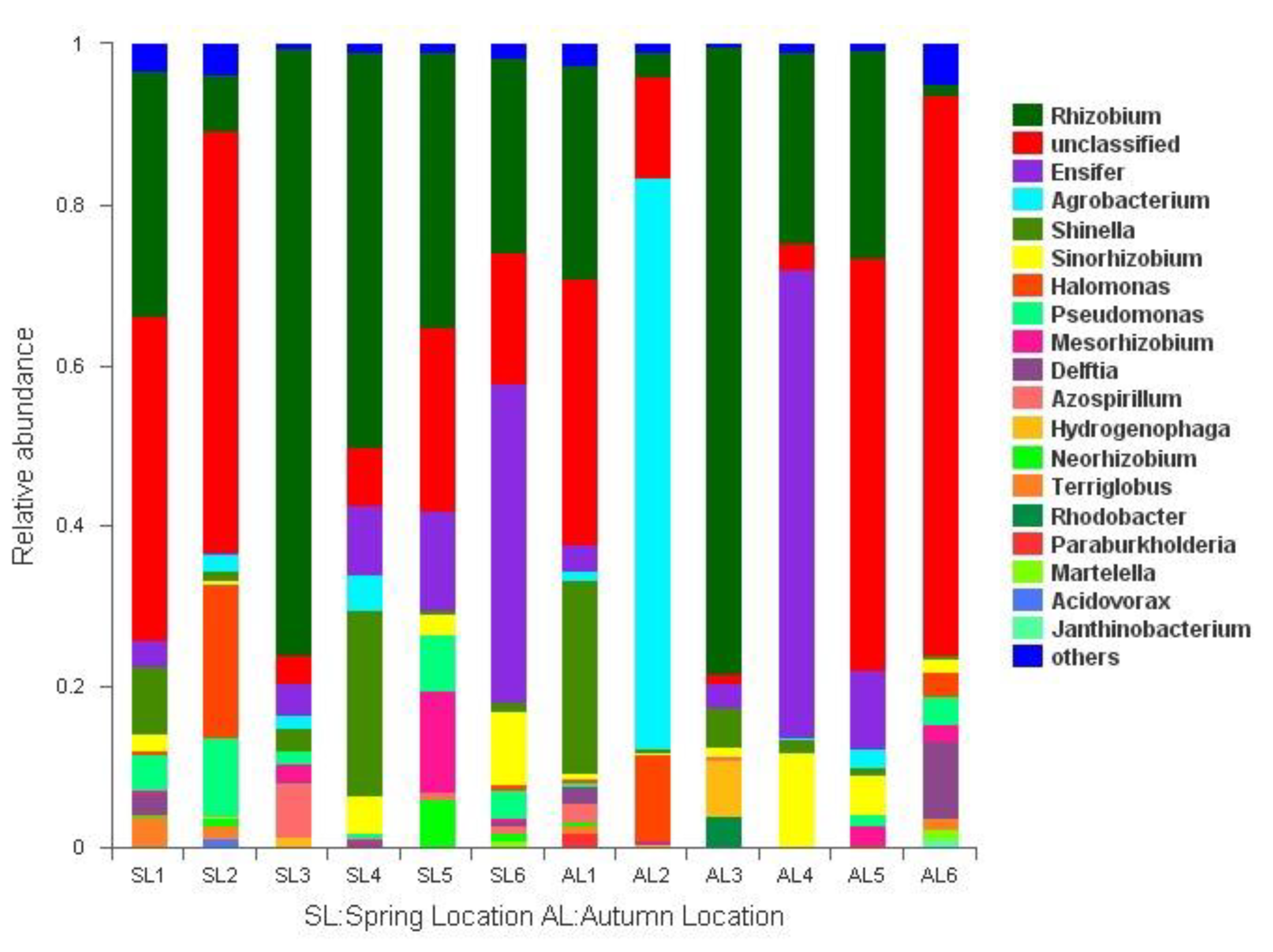
3.3. Changes in Composition of gcd-Harboring Bacterial Communities
3.3.1. Comparison of gcd-Harboring Bacterial Community Composition Based on Classification Level
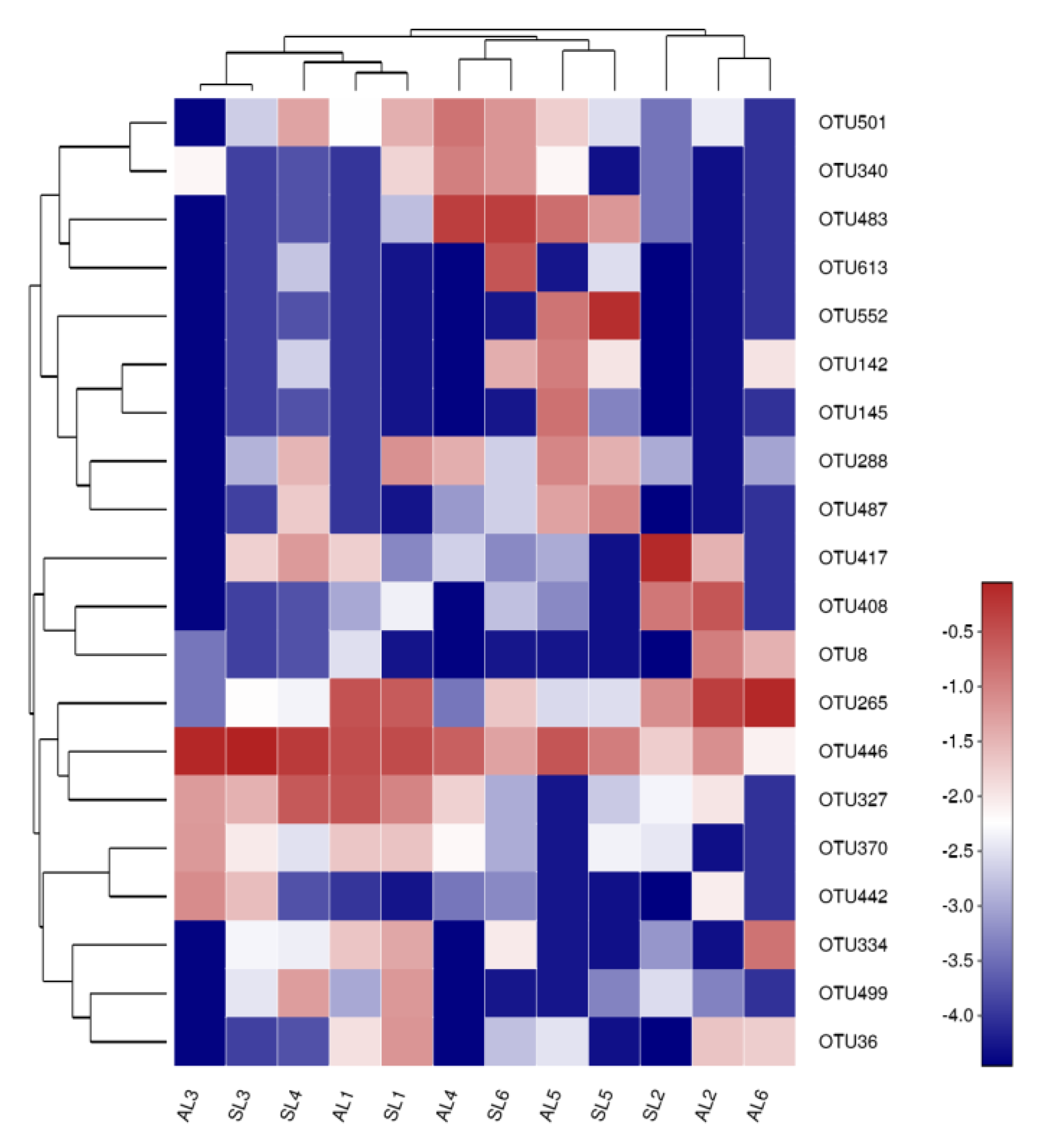
3.3.2. Analysis of the Composition of gcd-Harboring Bacterial Communities Based on OTU Level
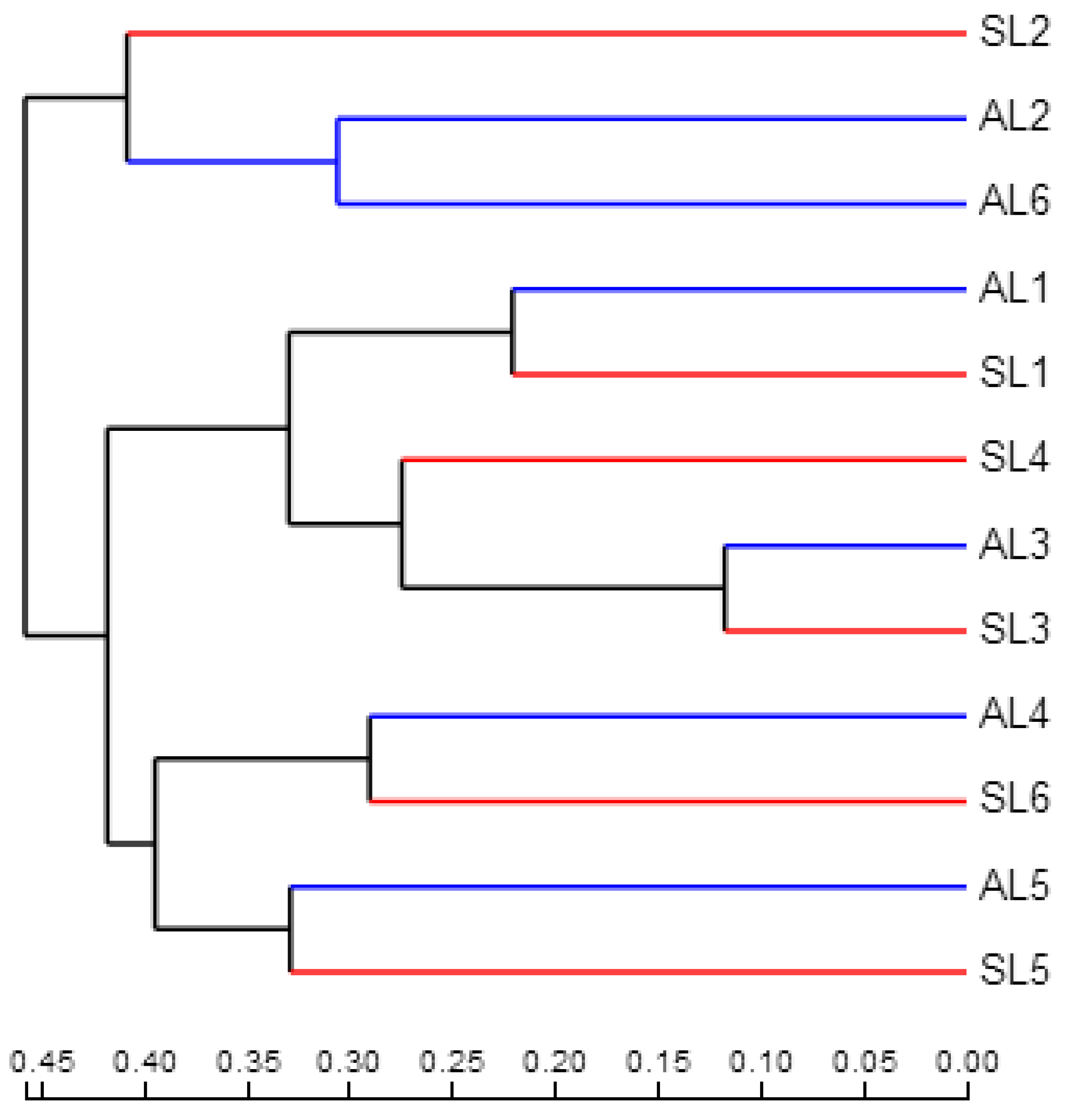
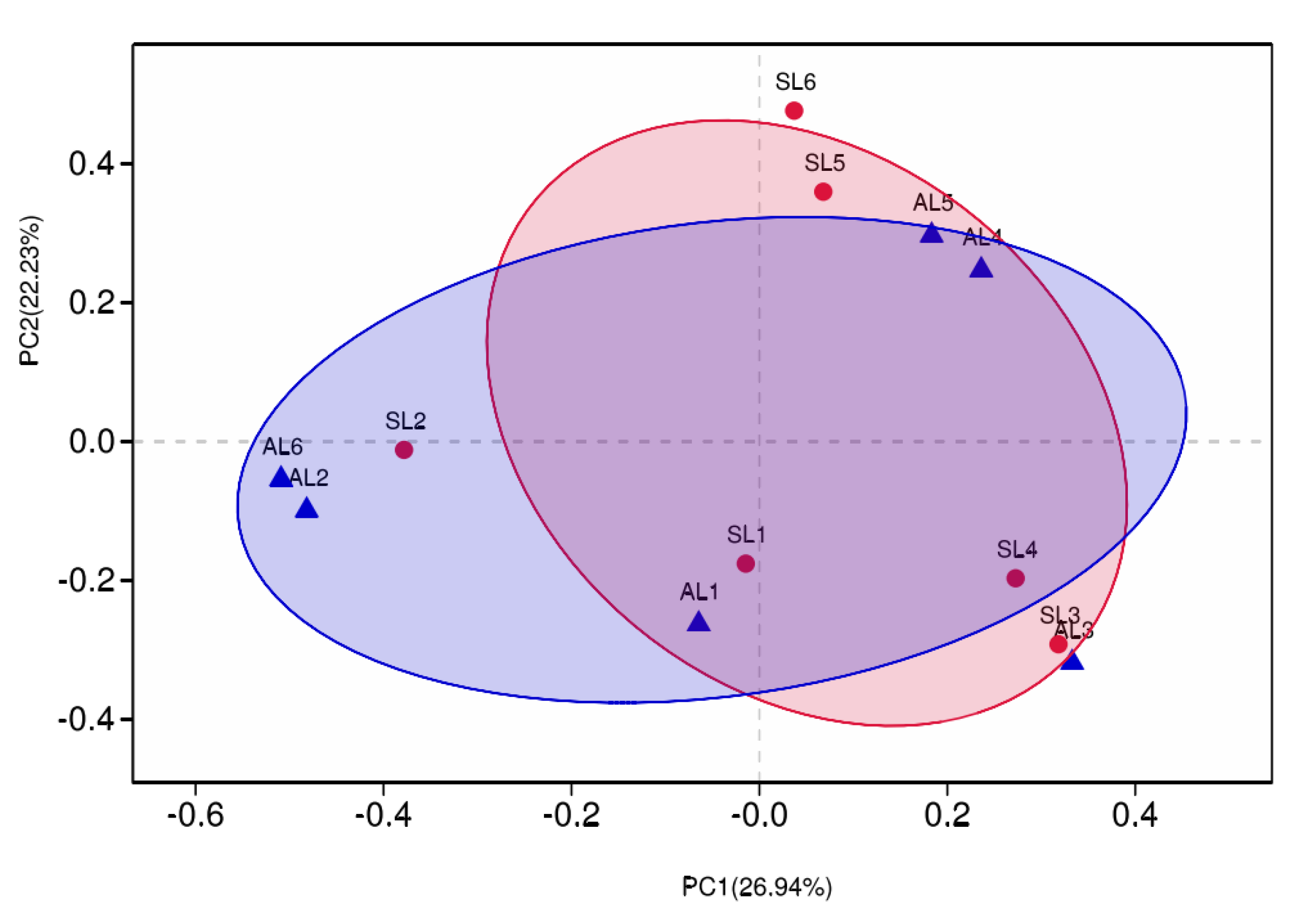
3.3.3. Comparative Analysis of gcd-Harboring Bacterial Community Composition Based on Beta Diversity
3.4. Correlation between gcd-Harboring Bacterial Communities and Environmental Conditions in the Sediment of Sancha Lake
3.4.1. Linking gcd-Harboring Bacterial Abundance, Diversity, and Community Structure with Environmental Factors
3.4.2. Correlation between Spatial Distribution Characteristics of gcd-Harboring Bacterial Communities and Environmental Conditions
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nilanjan, M.; Sanjib, K.M.; Srikanta, S. Ecological significance and phosphorus release potential of phosphate solubilizing bacteria in freshwater ecosystems. Hydrobiologia 2015, 745, 69–83. [Google Scholar]
- Zhao, X.R.; Lin, Q.M.; Li, B.G. Effect of C, N sources and C/N ratio on the solubilization of rock phosphate by some microorganisms. Plant Nutr. Fertil. Sci. 2002, 8, 197–204. [Google Scholar]
- Sashidhar, B.; Podile, A.R. Mineral phosphate solubilization by rhizosphere bacteriaand scope for manipulation of the direct oxidation pathwayinvolving glucose dehydrogenase. J. Appl. Microbiol. 2010, 109, 1–12. [Google Scholar]
- Goldstein, A.H.; Liu, S.T. Molecular cloning and regulation of a mineral phosphate solubilizing gene from Erwinia herbicola. Biotechnology 1987, 5, 72–74. [Google Scholar] [CrossRef]
- Cleton-Jansen, A.M.; Goosen, N.; Fayet, O. Cloning, mapping, and sequencing of the gene encoding Escherichia coli quinoprotein glucose dehydrogenase. J. Bacteriol. 1990, 172, 6308–6315. [Google Scholar] [CrossRef] [PubMed]
- Triipura, C.; Reddy, P.S.; Reddy, M.K. Glucose dehydrogenase of a rhizobacterial strain of Enterobacter asburiae involved in mineal phosphate solubilization shares properties and sequence homology with other members of enterobacteriaceae. Indian J. Microbiol. 2007, 47, 126–131. [Google Scholar] [CrossRef]
- Zeng, Q.W.; Wu, X.Q.; Wen, X.Y. Effects of Soluble Phosphate on Phosphate-Solubilizing Characteristics and Expression of gcd Gene in Pseudomonas frederiksbergensis JW-SD2. Curr. Microbiol. 2016, 72, 198–206. [Google Scholar] [CrossRef]
- Yang, P.X.; Ma, L.; Chen, M.H. Phosphate solubilizing ability and phylogenetic diversity of bacteria from p-rich soils around Dianchi Lake drainage area of China. Pedosphere 2012, 22, 707–716. [Google Scholar] [CrossRef]
- Bergkemper, F.; Kublik, S.; Lang, F. Novel oligonucleotide primers reveal a high diversity of microbes which drive phosphorous turnover in soil. J. Microbiol. Methods 2016, 125, 91–97. [Google Scholar] [CrossRef]
- Jia, B.Y.; Tang, Y.; Fu, W.L. Relationship among sediment characteristics, eutrophication process and human activities in the Sancha Lake. China Environ. Sci. 2013, 33, 1638–1644. [Google Scholar]
- Liu, J.W.; Yang, H.Z.; Zhao, M.X. Spatial distribution patterns of benthic microbial communities along the Pearl Estuary, China. Syst. Appl. Microbiol. 2014, 37, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Ruban, V.; Brigault, S.; Demare, D. An investigation of the origin and mobility of phosphorus in freshwater sediments from Bort-Les-Orgues Reservoir, France. J. Environ. Monit. 1999, 1, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Ruban, V.; López-Sánchez, J.F.; Pardo, P. Harmonized Protocol and certified reference material for the determination of extractable contents of phosphorus in freshwaer sediments—A synthesis of recent works. Fresenius J. Anal. Chem. 2001, 370, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.H.; Wang, D.H.; Wang, C.X. Relationship between phosphorus morphology in sediment and lake eutrophication. China Environ. Sci. 2003, 23, 583–586. [Google Scholar]
- General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China (AQSIQ). Determination of Total Organic Carbon in Sedimentary Rock; GB/T19145-2003; China Standards Press: Beijing, China, 2003. [Google Scholar]
- The Ministry of Environmental Protection of the People’s Republic of China. Determination of Total Nitrogen in Water-Alkaline Potassium Persulfate Digestion UV Spectrophotometry; HJ636-2012; China Standards Press: Beijing, China, 2012. [Google Scholar]
- The Ministry of Environmental Protection of the People’s Republic of China. Potassium Chloride Solution-Spectrophotometric Method; HJ634-2012; China Standards Press: Beijing, China, 2012. [Google Scholar]
- Chen, B.; The, B.S.; Sun, C. Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 2016, 6, 29505. [Google Scholar] [CrossRef]
- Ye, J.; Joseph, S.D.; Ji, M. Chemolithotrophic processes in the bacterial communities on the surface of mineral-enriched biochars. ISME J. 2017, 11, 1087–1101. [Google Scholar] [CrossRef]
- Calderón, K.; Spor, A.; Breuil, M.C. Effectiveness of ecological rescue for altered soil microbial communities and functions. ISME J. 2017, 11, 272–283. [Google Scholar] [CrossRef]
- Fan, X.; Xing, P. The vertical distribution of sediment archaeal community in the “black bloom” disturbing Zhushan Bay of Lake Taihu. Archaea 2016, 2016. [Google Scholar] [CrossRef]
- Mar, J.S.; Lamere, B.J.; Lin, D.L. Disease severity and immune activity relate to distinct interkingdom gut microbiome states in ethnically distinct ulcerative colitis patients. mBio 2016, 7, e01072-16. [Google Scholar] [CrossRef]
- Hu, H.; Chen, X.; Hou, F. Bacterial and fungal community structures in loess plateau grasslands with different grazing intensities. Front. Microbial. 2017, 8, 606. [Google Scholar]
- Rodriguez, H.; Fraga, R. Phosphatesolubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]




| Sample Site | Geographical Coordinates | Depth (m) | Hydrophyte | Description |
|---|---|---|---|---|
| L1 | 30°14′52″ N | 13 | Large quantity | Concentrated area of fenced breeding |
| 104°16′15″ E | ||||
| L2 | 30°14′28″ N | 4 | Large quantity | Tail water area of the reservoir |
| 104°15′32″ E | ||||
| L3 | 30°17′25″ N | 26 | Small quantity | Relatively concentrated area of fenced breeding |
| 104°16′31″ E | ||||
| L4 | 30°18′15″ N | 17 | Large quantity | Area with intense human activity |
| 104°14′31″ E | ||||
| L5 | 30°18′18″ N | 30 | Small quantity | Dense area of cage breeding |
| 104°16′2″ E | ||||
| L6 | 30°19′15″ N | 19 | Moderate quantity | Main water entry area of the lake |
| 104°15′14″ E |
| Season | Location | pH | DO (mg·L−1) | T (°C) | DTP (mg·L−1) | TOC (mg·g−1) | TN (mg·g−1) | NH3-N (mg·g−1) | TP (mg·g−1) | OP | IP (mg·g−1) | HCl–P (mg·g−1) | NaOH–P (mg·g−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spring | L1 | 7.38 ± 0.11 | 6.4 ± 1.0 | 13.0 ± 0.3 | 0.088 ± 0.025 | 48.1 ± 5.0 | 6.46 ± 0.46 | 0.387 ± 0.030 | 1.036 ± 0.100 | 0.242 ± 0.042 | 0.790 ± 0.090 | 0.590 ± 0.01 | 0.148 ± 0.008 |
| L2 | 7.52 ± 0.13 | 6.4 ± 0.9 | 12.9 ± 0.1 | 0.092 ± 0.040 | 36.9 ± 4.0 | 4.30 ± 0.30 | 0.057 ± 0.007 | 0.715 ± 0.015 | 0.229 ± 0.090 | 0.617 ± 0.017 | 0.550 ± 0.010 | 0.047 ± 0.010 | |
| L3 | 7.52 ± 0.10 | 5.8 ± 0.2 | 12.6 ± 0.3 | 0.035 ± 0.005 | 76.6 ± 8.0 | 10.15 ± 1.00 | 0.017 ± 0.007 | 3.069 ± 0.092 | 0.562 ± 0.062 | 2.687 ± 0.087 | 2.248 ± 0.100 | 0.367 ± 0.060 | |
| L4 | 7.46 ± 0.10 | 6.7 ± 1.1 | 13.2 ± 0.2 | 0.069 ± 0.002 | 55.0 ± 6.0 | 4.87 ± 0.87 | 0.021 ± 0.000 | 1.120 ± 0.120 | 0.189 ± 0.046 | 0.824 ± 0.004 | 0.786 ± 0.050 | 0.099 ± 0.009 | |
| L5 | 7.45 ± 0.09 | 5.1 ± 0.5 | 12.6 ± 0.2 | 0.033 ± 0.003 | 55.6 ± 5.0 | 6.57 ± 0.50 | 0.067 ± 0.007 | 1.376 ± 0.109 | 0.311 ± 0.011 | 1.162 ± 0.10 | 0.556 ± 0.010 | 0.340 ± 0.020 | |
| L6 | 7.67 ± 0.21 | 9.0 ± 1.0 | 12.7 ± 0.3 | 0.065 ± 0.005 | 25.4 ± 3.0 | 1.66 ± 0.06 | 0.096 ± 0.006 | 0.696 ± 0.100 | 0.099 ± 0.009 | 0.481 ± 0.090 | 0.406 ± 0.006 | 0.077 ± 0.007 | |
| Autumn | L1 | 7.54 ± 0.10 | 5.6 ± 0.1 | 15.4 ± 0.4 | 0.057 ± 0.007 | 47.5 ± 2.0 | 4.49 ± 0.49 | 0.244 ± 0.034 | 2.874 ± 0.074 | 0.317 ± 0.017 | 2.603 ± 0.060 | 1.088 ± 0.088 | 0.172 ± 0.002 |
| L2 | 6.89 ± 0.19 | 5.5 ± 0.1 | 17.2 ± 1.0 | 0.051 ± 0.002 | 34.8 ± 1.8 | 3.18 ± 0.28 | 0.035 ± 0.005 | 1.153 ± 0.053 | 0.444 ± 0.021 | 0.782 ± 0.030 | 0.450 ± 0.050 | 0.395 ± 0.005 | |
| L3 | 7.11 ± 0.10 | 4.1 ± 0.1 | 14.6 ± 0.6 | 0.012 ± 0.001 | 83.2 ± 3.2 | 10.77 ± 1.74 | 0.539 ± 0.039 | 5.005 ± 0.202 | 0.609 ± 0.041 | 3.882 ± 0.080 | 3.517 ± 0.017 | 1.007 ± 0.007 | |
| L4 | 7.16 ± 0.06 | 5.5 ± 0.2 | 16.3 ± 0.9 | 0.055 ± 0.005 | 54.2 ± 2.4 | 3.99 ± 0.41 | 0.141 ± 0.004 | 2.661 ± 0.060 | 0.295 ± 0.010 | 1.897 ± 0.070 | 1.949 ± 0.049 | 0.222 ± 0.022 | |
| L5 | 7.02 ± 0.02 | 4.5 ± 0.1 | 14.1 ± 0.1 | 0.015 ± 0.005 | 47.8 ± 2.0 | 5.25 ± 0.79 | 0.071 ± 0.001 | 1.454 ± 0.054 | 0.369 ± 0.009 | 1.051 ± 0.051 | 0.554 ± 0.054 | 0.611 ± 0.011 | |
| L6 | 7.18 ± 0.18 | 5.9 ± 0.3 | 16.5 ± 0.5 | 0.025 ± 0.005 | 32.9 ± 1.7 | 2.28 ± 0.18 | 0.265 ± 0.006 | 0.629 ± 0.029 | 0.167 ± 0.007 | 0.444 ± 0.024 | 0.343 ± 0.033 | 0.104 ± 0.004 |
| Season | Sampling Site | Reads | Chao1 | Shannon | Coverage | OTUs | No. of Phyla | No. of Classes | No. of Orders | No. of Families | No. of Genera |
|---|---|---|---|---|---|---|---|---|---|---|---|
| spring | L1 | 32,585 ± 150 | 234.55 ± 33.05 | 3.365 ± 0.040 | 0.9993 ± 0.0004 | 226 ± 6 | 6 | 9 | 13 | 19 | 22 |
| L2 | 23,251 ± 120 | 156.18 ± 21.04 | 2.777 ± 0.067 | 0.9985 ± 0.0012 | 150 ± 4 | 5 | 9 | 12 | 15 | 23 | |
| L3 | 12,834 ± 60 | 98.60 ± 19.04 | 1.539 ± 0.030 | 0.9986 ± 0.0007 | 85 ± 2 | 2 | 4 | 7 | 11 | 16 | |
| L4 | 41,828 ± 200 | 185.27 ± 29.04 | 2.622 ± 0.064 | 0.9993 ± 0.0005 | 158 ± 4 | 1 | 3 | 5 | 10 | 17 | |
| L5 | 12,151 ± 60 | 140.69 ± 18.09 | 3.167 ± 0.071 | 0.9983 ± 0.0002 | 130 ± 3 | 3 | 5 | 7 | 10 | 13 | |
| L6 | 13,357 ± 65 | 322.14 ± 36.64 | 3.397 ± 0.075 | 0.9958 ± 0.0017 | 291 ± 7 | 3 | 6 | 9 | 17 | 32 | |
| autumn | L1 | 95,695 ± 50 | 163.05 ± 23.04 | 2.943 ± 0.040 | 0.9978 ± 0.0014 | 154 ± 4 | 5 | 8 | 12 | 16 | 22 |
| L2 | 10,880 ± 54 | 143.96 ± 20.05 | 1.259 ± 0.014 | 0.9983 ± 0.0004 | 121 ± 3 | 2 | 4 | 5 | 5 | 15 | |
| L3 | 3199 ± 10 | 43.00 ± 1.00 | 1.307 ± 0.016 | 0.9974 ± 0.0000 | 29 ± 0 | 1 | 3 | 5 | 6 | 10 | |
| L4 | 10,852 ± 54 | 80.20 ± 3.04 | 1.939 ± 0.029 | 0.9984 ± 0.0006 | 62 ± 2 | 1 | 3 | 5 | 7 | 10 | |
| L5 | 30,857 ± 150 | 139.38 ± 17.04 | 2.432 ± 0.039 | 0.9994 ± 0.0001 | 118 ± 3 | 2 | 3 | 5 | 8 | 12 | |
| L6 | 18,415 ± 92 | 157.53 ± 22.04 | 2.273 ± 0.027 | 0.9992 ± 0.0005 | 152 ± 3 | 5 | 8 | 13 | 21 | 23 |
| Environmental Factor | PH | DO | T | TN | TOC | TP | HCl-P | NaOH-P | DTP |
|---|---|---|---|---|---|---|---|---|---|
| OTUs | 0.422 a/0.172 b | 0.346/0.271 | 0.239/0.454 | –0.577/0.05 * | –0.631/0.028 * | –0.698/0.012 * | –0.687/0.014 * | –0.624/0.03 * | 0.915/0.000 ** |
| Chao1 | 0.043/0.894 | 0.391/0.211 | –0.063/0.846 | –0.604/0.037 * | –0.6/0.039 * | –0.619/0.032 * | –0.653/0.021 * | –0.32/0.311 | 0.643/0.024 * |
| Shannon | 479/0.115 | 0.251/0.432 | 0.278/0.382 | –0.574/0.051 | –0.619/0.032 * | –702/0.011 * | –0.672/0.017 * | –0.635/0.26 | 0.921/0.000 ** |
| No. of filtered reads | 0.371/0.236 | 0.907/0.002 ** | 0.12/0.711 | –0.093/0.773 | –0.191/0.553 | –0.464/0.129 | –0.457/0.135 | –514/0.088 | 0.602/0.039 * |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhang, J.; Gong, Z.; Xu, W.; Mou, Z. Gcd Gene Diversity of Quinoprotein Glucose Dehydrogenase in the Sediment of Sancha Lake and Its Response to the Environment. Int. J. Environ. Res. Public Health 2019, 16, 1. https://doi.org/10.3390/ijerph16010001
Li Y, Zhang J, Gong Z, Xu W, Mou Z. Gcd Gene Diversity of Quinoprotein Glucose Dehydrogenase in the Sediment of Sancha Lake and Its Response to the Environment. International Journal of Environmental Research and Public Health. 2019; 16(1):1. https://doi.org/10.3390/ijerph16010001
Chicago/Turabian StyleLi, Yong, Jianqiang Zhang, Zhiliang Gong, Wenlai Xu, and Zishen Mou. 2019. "Gcd Gene Diversity of Quinoprotein Glucose Dehydrogenase in the Sediment of Sancha Lake and Its Response to the Environment" International Journal of Environmental Research and Public Health 16, no. 1: 1. https://doi.org/10.3390/ijerph16010001




