Dynamics of Bacterial and Fungal Communities during the Outbreak and Decline of an Algal Bloom in a Drinking Water Reservoir
Abstract
:1. Introduction
2. Experimental Section
2.1. Study Site and Sample Collection
2.2. Water Physical and Chemical Parameters Measurement
2.3. Algal Cell Concentration and Biomass Determination
2.4. Water Microbial DNA Extraction
2.5. Illumina MiSeq Sequencing and Sequence Analysis
2.6. Nucleotide Sequence Accession Number
2.7. Statistical Analysis
3. Results and Discussion
3.1. Water Quality Parameters
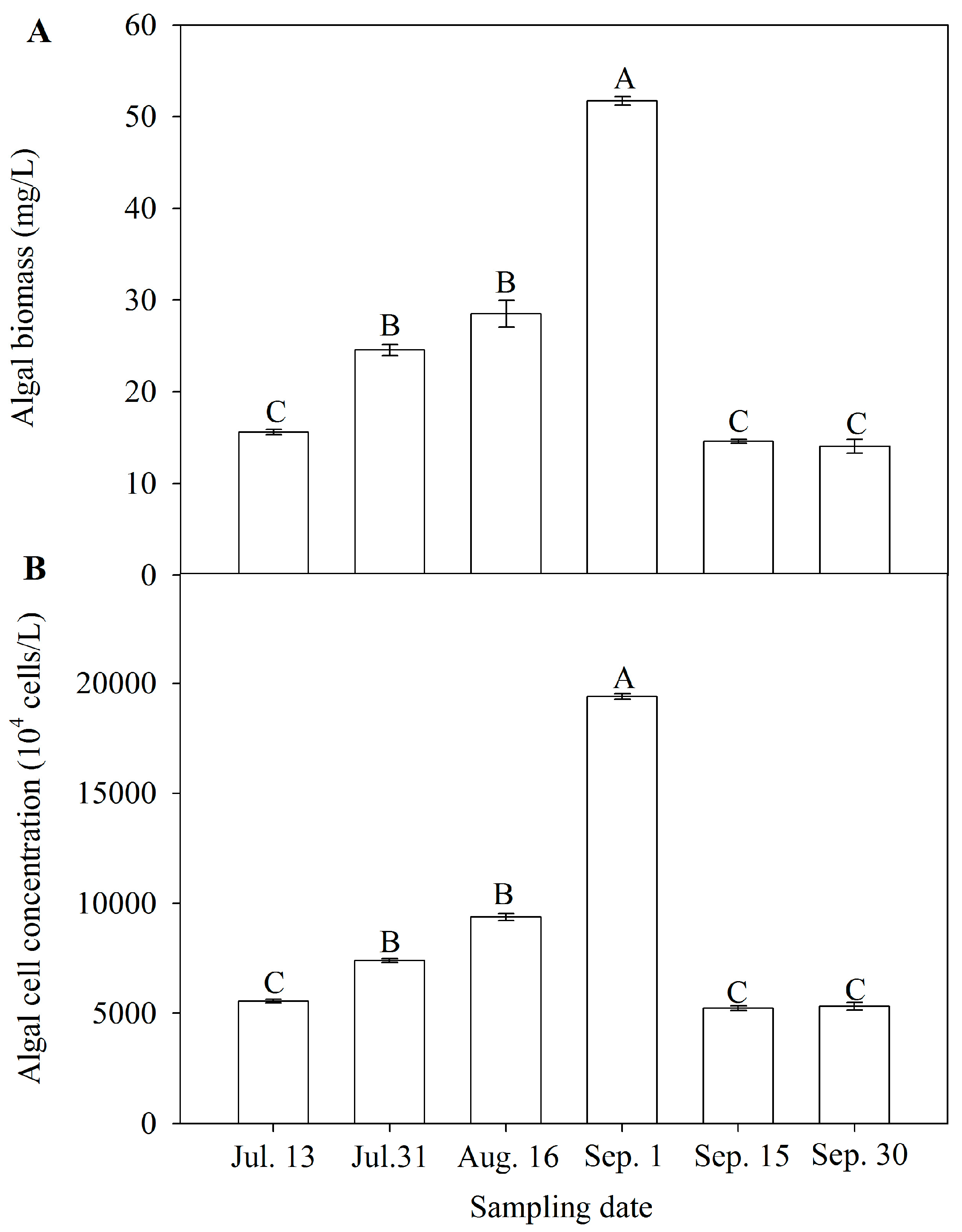
3.2. Algal Biomass and Cell Concentration
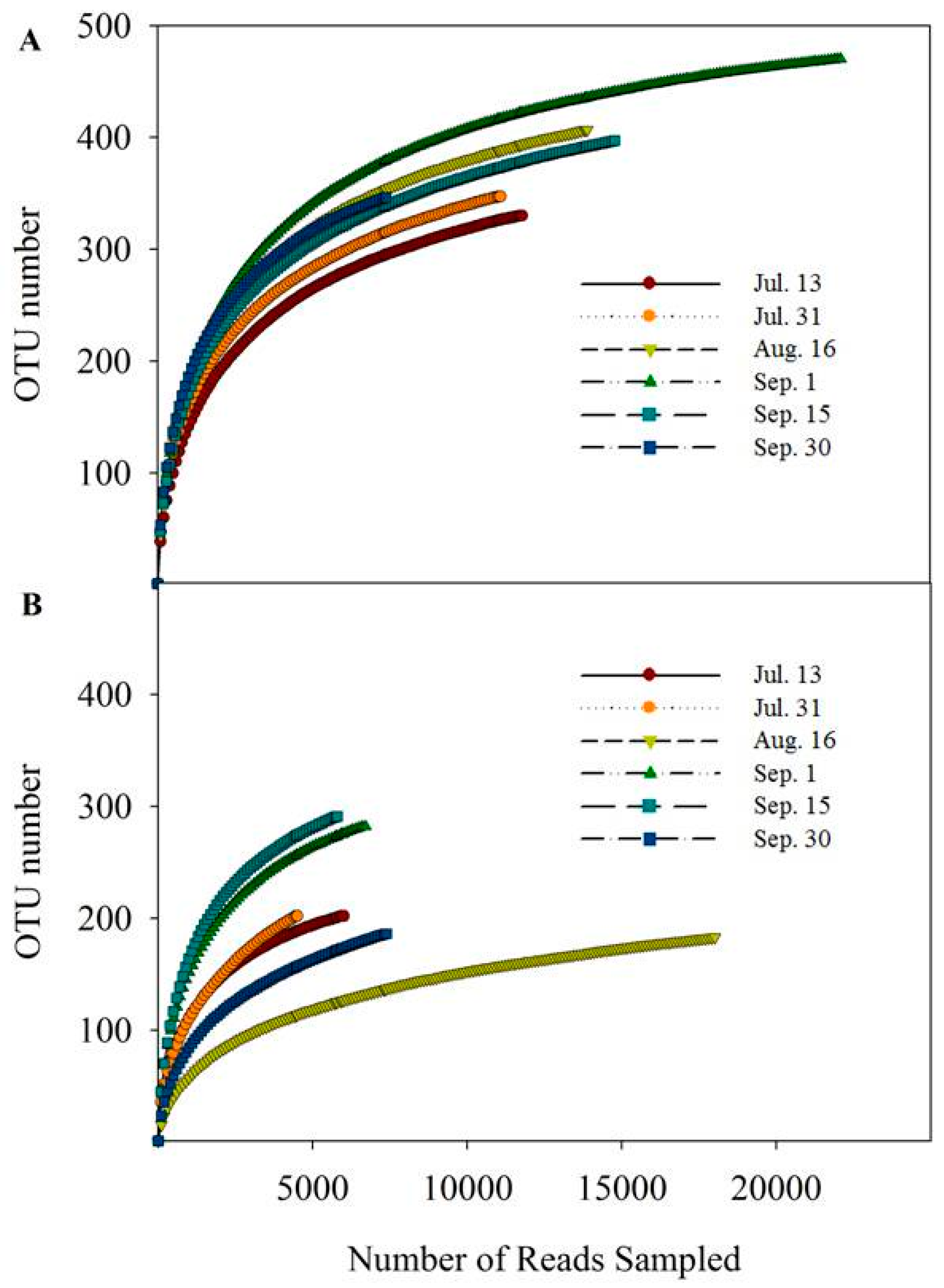
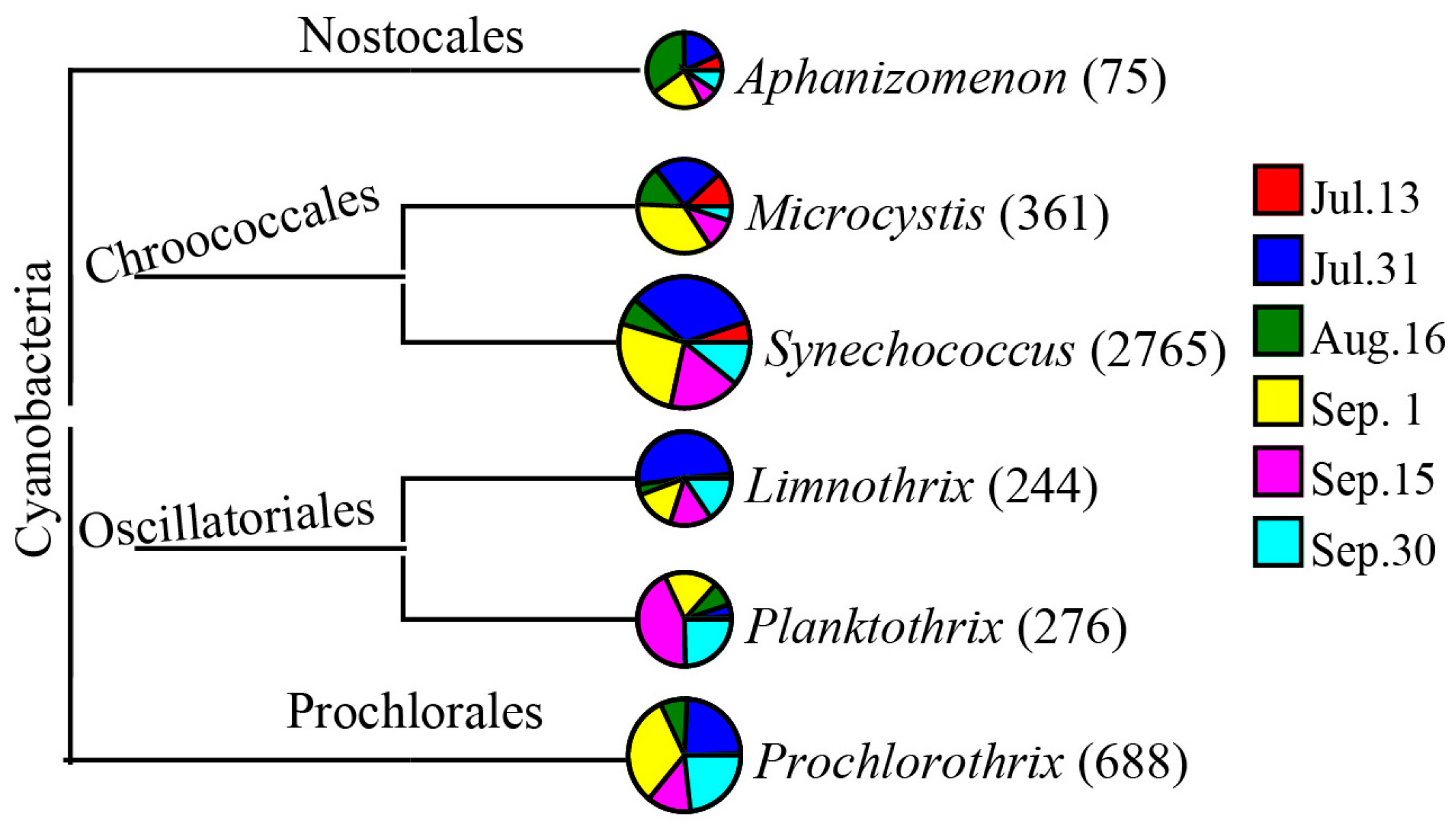
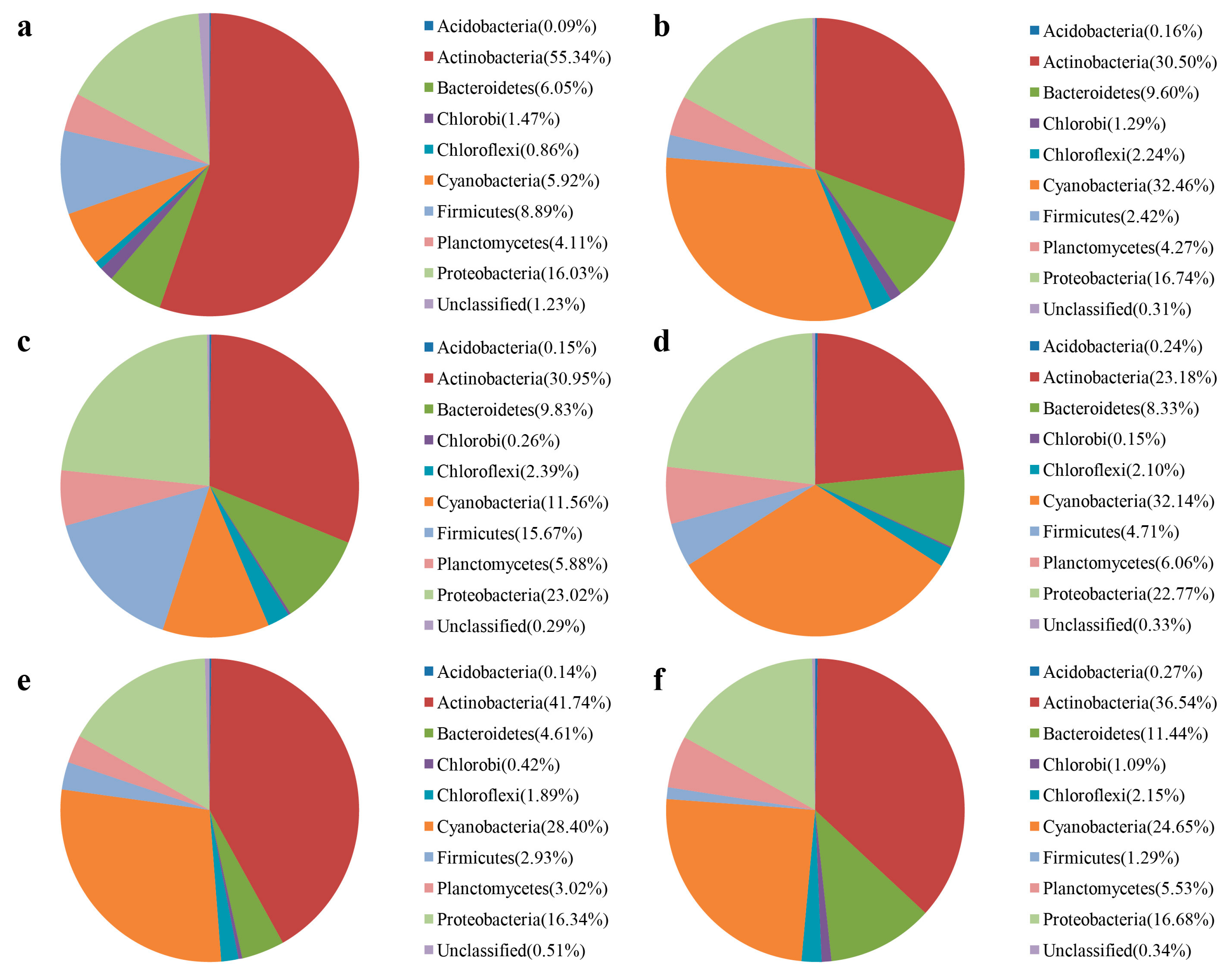
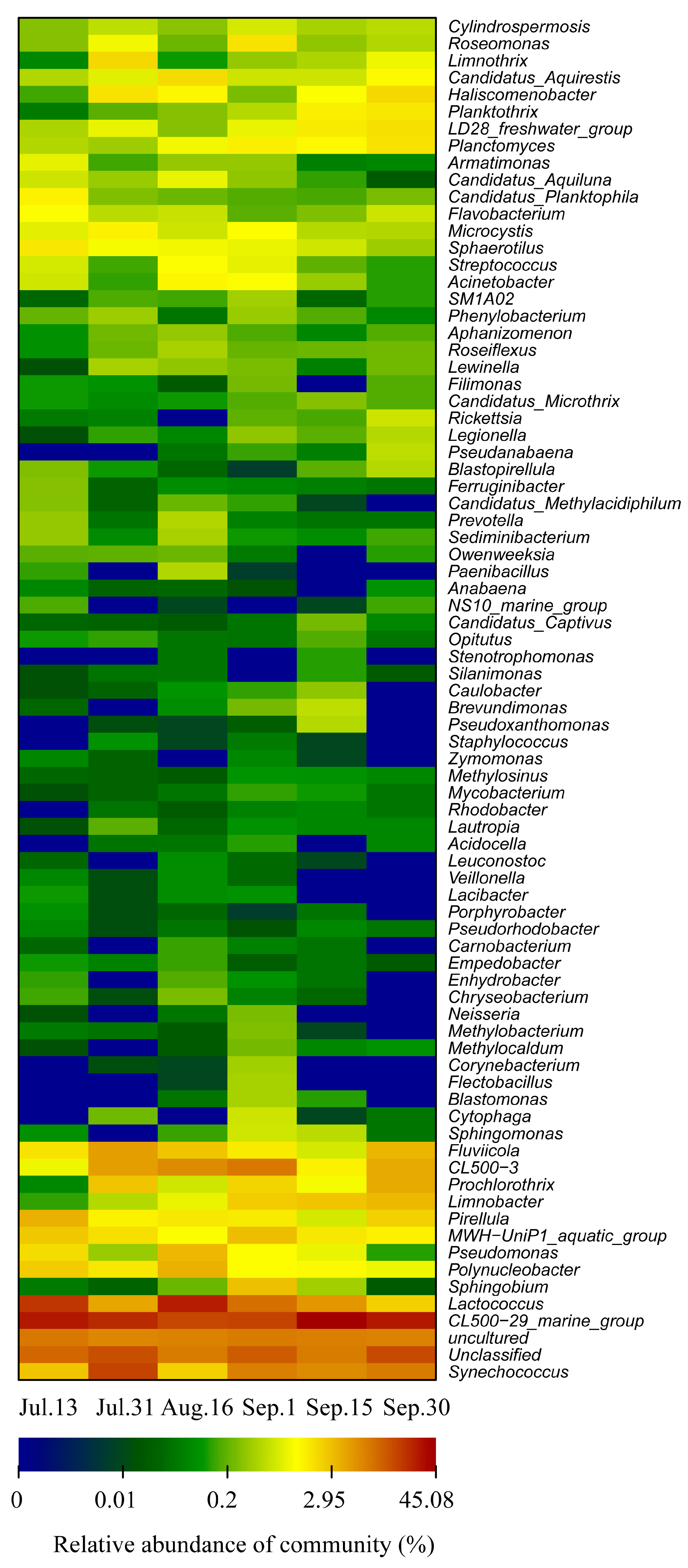
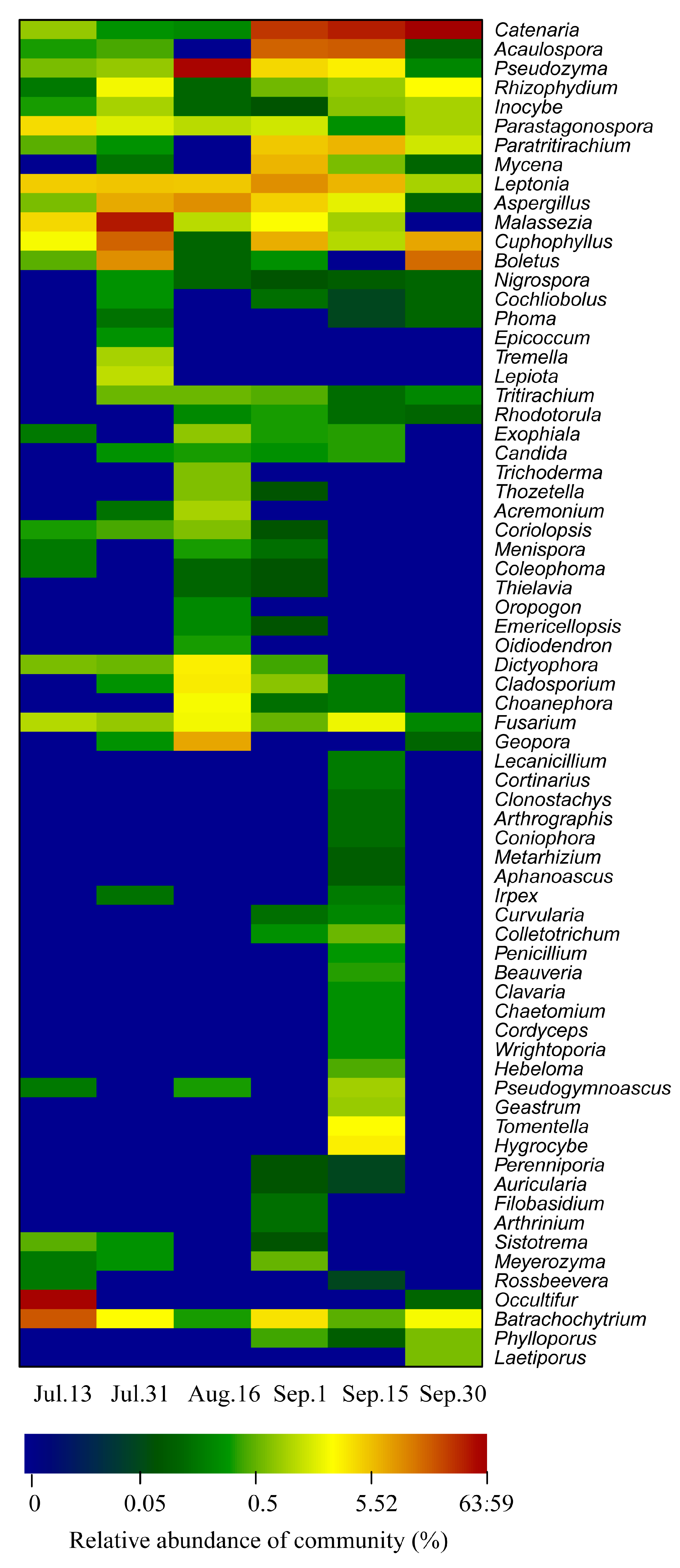
3.3. Water Bacterial and Fungal Communities
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Šulčius, S.; Montvydienė, D.; Mazur-Marzec, H.; Kasperovičienė, J.; Rulevičius, R.; Cibulskaitė, Z. The profound effect of harmful cyanobacterial blooms: From food-web and management perspectives. Sci. Total Environ. 2017, 609, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Hall, N.S.; Calandrino, E.S. Controlling harmful cyanobacterial blooms in a world experiencing anthropogenic and climatic-induced change. Sci. Total Environ. 2011, 409, 1739–1745. [Google Scholar] [CrossRef] [PubMed]
- Metson, G.S.; Lin, J.J.; Harrison, J.A.; Compton, J.E. Linking terrestrial phosphorus inputs to riverine export across the United States. Water Res. 2017, 124, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Jiang, Y.; Wang, Z.; Peng, L.; Luo, S.; Gu, J.; Li, R. Interactions between algicidal bacteria and the cyanobacterium Microcystis aeruginosa: Lytic characteristics and physiological responses in the cyanobacteria. Int. J. Environ. Sci. Technol. 2014, 11, 469–476. [Google Scholar] [CrossRef]
- Berry, M.A.; Davis, T.W.; Cory, R.M.; Duhaime, M.B.; Johengen, T.H.; Kling, G.W.; Marino, J.A.; Den Uyl, P.A.; Gossiaux, D.; Dick, G.J.; et al. Cyanobacterial harmful algal blooms are a biological disturbance to Western Lake Erie bacterial communities. Environ. Microbiol. 2017, 19, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Parulekar, N.N.; Kolekar, P.; Jenkins, A.; Kleiven, S.; Utkilen, H.; Johansen, A.; Sawant, S.; Kulkarni-Kale, U.; Kale, M.; Sæbø, M. Characterization of bacterial community associated with phytoplankton bloom in a eutrophic lake in South Norway using 16S rRNA gene amplicon sequence analysis. PLoS ONE 2017, 12, e0173408. [Google Scholar] [CrossRef] [PubMed]
- Scherer, P.I.; Millard, A.D.; Miller, A.; Schoen, R.; Raeder, U.; Geist, J.; Zwirglmaier, K. Temporal dynamics of the microbial community composition with a focus on toxic cyanobacteria and toxin presence during harmful algal blooms in two South German lakes. Front. Microbiol. 2017, 8, 2387. [Google Scholar] [CrossRef] [PubMed]
- Su, X.M.; Steinman, A.D.; Tang, X.M.; Xue, Q.J.; Zhao, Y.Y.; Xie, L.Q. Response of bacterial communities to cyanobacterial harmful algal blooms in Lake Taihu, China. Harmful Algae. 2017, 68, 168–177. [Google Scholar] [CrossRef] [PubMed]
- González, J.M.; Simó, R.; Massana, R.; Covert, J.S.; Casamayor, E.O.; Pedrós-Alió, C.; Moran, M.A. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 2000, 66, 4237–4246. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Huang, T.L.; Zhang, H.H. Effects of seasonal thermal stratification on the functional diversity and composition of the microbial community in a drinking water reservoir. Water 2015, 7, 5525–5546. [Google Scholar] [CrossRef]
- Zhang, H.H.; Huang, T.L.; Chen, S.N.; Yang, X.; Lv, K.; Raju, S. Abundance and diversity of bacteria in oxygen minimum drinking water reservoir sediments studied by quantitative PCR and Pyrosequencing. Microb. Ecol. 2015, 69, 618–629. [Google Scholar]
- Zhang, H.H.; Huang, T.L.; Chen, S.N. Ignored sediment fungal populations in water supply reservoirs are revealed by quantitative PCR and 454 Pyrosequencing. BMC Microbiol. 2015, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.H.; Huang, T.L.; Chen, S.N.; Guo, L.; Liu, T.T.; Yang, X. Microbial community functional diversity and enzymatic activity in the sediments of drinking water reservoirs, Northwest China. Desalin. Water Treat. 2014, 52, 1608–1614. [Google Scholar] [CrossRef]
- Jia, Y.; Du, J.J.; Fang, H.; Zhao, G.Y.; Tian, X.J. Inhibition of freshwater algal species by co-culture with two fungi. Mater. Sci. Eng. 2013, 33, 2451–2454. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.P.; Huang, T.L.; Zeng, M.Z. Differences in phytoplankton dynamics and community Structure between a wet year and dry year in the Zhoucun Reservoir. J. Freshwater Ecol. 2016, 31, 377–391. [Google Scholar] [CrossRef]
- Shi, J.C.; Huang, T.L.; Wen, G.; Liu, F.; Qiu, X.P.; Wang, B.S. The variation characteristic of sulfides and VOSc in a source water reservoir and its control using a water-lifting aerator. Int. J. Environ. Res. Public Health 2016, 13, 427. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.E.; Clesceri, L.S.; Eaton, A.D. Standard Methods for the Examination of Water and Wastewater, 18th ed.; American Public Health Association: Washington, DC, USA, 1992. [Google Scholar]
- Nazeer, M.; Wong, M.S.; Nichol, J.E. A new approach for the estimation of phytoplankton cell counts associated with algal blooms. Sci. Total Environ. 2017, 590, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.S.; He, S.M.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.L.; Tringe, S.G. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015, 6, 771. [Google Scholar] [CrossRef] [PubMed]
- Degnan, P.H.; Ochman, H. Illumina-based analysis of microbial community diversity. ISME J. 2012, 6, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinformatics 2011, 1E–5E. [Google Scholar] [CrossRef]
- Sun, W.M.; Xiao, E.Z.; Xiao, T.F.; Krumins, V.; Wang, Q.; Häggblom, M.; Dong, Y.R.; Tang, S.; Hu, M.; Li, B.Q. Response of soil microbial communities to elevated antimony and arsenic contamination indicates the relationship between the innate microbiota and contaminant fractions. Environ. Sci. Technol. 2017, 51, 9165–9175. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing MOTHUR: Open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.Y.; Wu, W.M.; Wu, L.Y.; He, Z.L.; Nostrand, J.D.V.; Deng, Y.; Luo, J.; Carley, J.; Ginder-Vogel, M.; Gentry, T.J.; et al. Responses of microbial community functional structures to pilot-scale uranium in situ bioremediation. ISME J. 2010, 4, 1060–1070. [Google Scholar] [CrossRef] [PubMed]
- Su, X.M.; Steinman, A.D.; Xue, Q.J.; Zhao, Y.Y.; Tang, X.M.; Xie, L.Q. Temporal patterns of phyto- and bacterioplankton and their relationships with environmental factors in Lake Taihu, China. Chemosphere 2017, 184, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Sison-Mangus, M.P.; Jiang, S.; Kudela, R.M.; Mehic, S.J. Phytoplankton-associated bacterial community composition and succession during toxic diatom bloom and non-bloom events. Front. Microbiol. 2016, 7, 1433. [Google Scholar] [CrossRef] [PubMed]
- Luria, C.M.; Amaral-Zettler, L.A.; Ducklow, H.W.; Repeta, D.J.; Rhyne, A.L.; Rich, J.J. Seasonal shifts in bacterial community responses to phytoplankton-derived dissolved organic matter in the Western Antarctic Peninsula. Front. Microbiol. 2017, 8, 2117. [Google Scholar] [CrossRef] [PubMed]
- Landa, M.; Blain, S.; Christaki, U.; Monchy, S.; Obernosterer, I. Shifts in bacterial community composition associated with increased carbon cycling in a mosaic of phytoplankton blooms. ISME J. 2016, 10, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.Y.; Cao, X.Y.; Huang, R.; Zeng, J.; Wu, Q.L.L. Variation of bacterial communities in water and sediments during the decomposition of Microcystis biomass. PLoS ONE 2017, 12, e0176397. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.Y.; Yu, Z.; Chen, H.H.; Yang, J.R.; Liu, M.; Liu, L.M.; Huang, B.Q.; Yang, J. Cyanobacterial bloom significantly boosts hypolimnelic anammox bacterial abundance in a subtropical stratified reservoir. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef] [PubMed]
- Olapade, O.A. Community composition and diversity of coastal Bacterioplankton assemblages in Lakes Michigan, Erie, and Huron. Microb. Ecol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Rinta-Kanto, J.M.; Sun, S.L.; Sharma, S.; Kiene, R.P.; Moran, M.A. Bacterial community transcription patterns during a marine phytoplankton bloom. Environ. Microbiol. 2012, 14, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Gallego, V.; Sánchez-Porro, C.; García, M.T.; Ventosa, A. Roseomonas aquatica sp. nov., isolated from drinking water. Int. J. Syst. Evol. Microbiol. 2006, 56, 2291–2295. [Google Scholar]
- Teeling, H.; Fuchs, B.M.; Becher, D.; Klockow, C.; Gardebrecht, A.; Bennke, C.M.; Kassabgy, M.; Huang, S.X.; Mann, A.J.; Waldmann, J.; et al. Substrate-controlled succession of marine Bacterioplankton populations induced by a phytoplankton bloom. Science 2012, 336, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Zeder, M.; Peter, S.; Shabarova, T.; Pernthaler, J. A small population of planktonic Flavobacteria with disproportionally high growth during the spring phytoplankton bloom in a prealpine lake. Environ. Microbiol. 2009, 11, 2676–2686. [Google Scholar] [CrossRef] [PubMed]
- Taube, R.; Ganzert, L.; Grossart, H.P.; Gleixner, G.; Premke, K. Organic matter quality structures benthic fatty acid patterns and the abundance of fungi and bacteria in temperate lakes. Sci. Total Environ. 2018, 610, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Y.; Song, Y.; Ma, Z.P.; Zhang, H.J.; Yang, Z.D.; Cai, Z.H.; Zhou, J. Fungal community dynamics during a marine dinoflagellate (Noctiluca scintillans) bloom. Mar. Environ. Res. 2017, 131, 183–194. [Google Scholar]
- Lefèvre, E.; Roussel, B.; Amblard, C.; Sime-Ngando, T. The molecular diversity of freshwater picoeukaryotes reveals high occurrence of putative parasitoids in the plankton. PLoS One 2008, 3, e2324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasconi, S.; Jobard, M.; Sime-Ngando, T. Parasitic fungi of phytoplankton: Ecological roles and implications for microbial food webs. Aquat. Microb. Ecol. 2011, 62, 123–137. [Google Scholar] [CrossRef]
- Zhang, W.G.; Gao, Y.; Yi, N.; Wang, C.C.; Di, P.P.; Yan, S.H. Variations in abundance and community composition of denitrifying bacteria during a cyanobacterial bloom in a eutrophic shallow lake in China. J. Freshwater Ecol. 2017, 32, 467–476. [Google Scholar] [CrossRef]
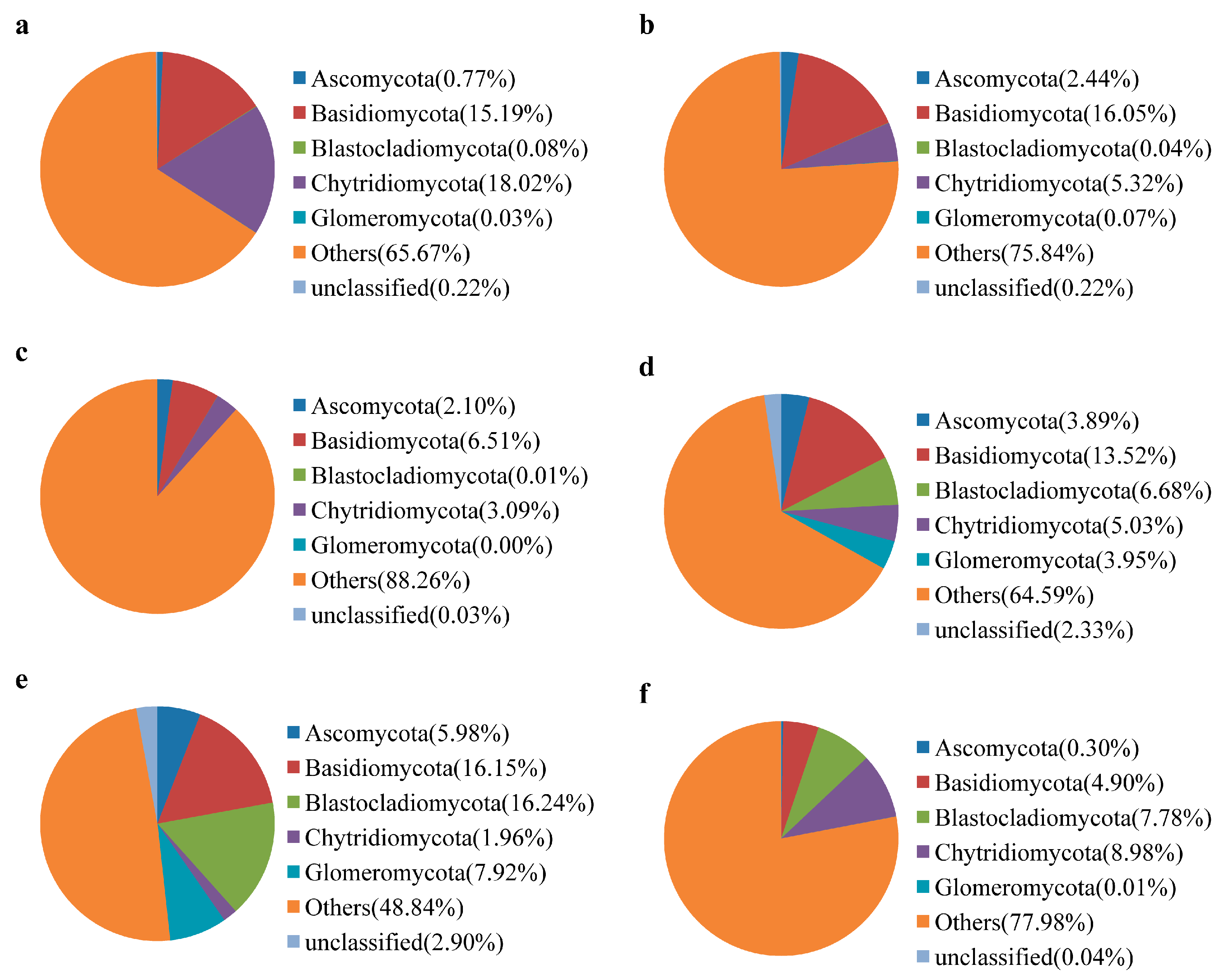

| Water Microbe | Amplified Nuclear Ribosomal Regions | Primer Sets (from 5' to 3') | PCR Program |
|---|---|---|---|
| Bacteria [19,20] | 16S rRNA V4-V5 region | 515F: GTGCCAGCMGCCGCGGTAA 907R: CCGTCAATTCCTTTGAGTTT | 95°C for 2 min, 30 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, with a final extension at 72°C for 5 min |
| Fungi [21] | Internal transcribed spacer-1 (ITS1) region | 1737F:GGAAGTAAAAGTCGTAACAAGG 2043R:GCTGCGTTCTTCATCGATGC | 95°C for 1 min, 30 cycles of 95°C for 10 s, 55°C for 30 s, 72°C for 30 s, with a final extension at 72°C for 5 min |
| Sampling date | T (℃) | DO (mg/L) | Turbidity (NTU) | Conductivity (S/cm) | pH | CODMn (mg/L) | TN (mg/L) | NO3−-N (mg/L) | NO2−-N (mg/L) | NH4+-N (mg/L) | TP (mg/L) | Fe (mg/L) | Mn (mg/L) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Jul. 13 | 28 ± 0.06 | 12.5 ± 0.12 | 7.3 ± 0.2 | 271 ± 1.7 | 8.38 ± 0.01 | 4.46 ± 0.28 | 1.80 ± 0.02 | 0.91 ± 0.01 | 0.03 ± 0.0 | 0.34 ± 0.03 | 0.07 ± 0.0 | 0.05 ± 0.0 | 0.08 ± 0.0 |
| Jul. 31 | 31 ± 0.06 | 7.3 ± 0.3 | 14.7 ± 0.1 | 286 ± 3.5 | 8.65 ± 0.02 | 7.52 ± 0.02 | 2.6 ± 0.03 | 1.44 ± 0.03 | 0.15 ± 0.0 | 0.45 ± 0.03 | 0.06 ± 0.0 | 0.11 ± 0.0 | 0.09 ± 0.0 |
| Aug. 16 | 29 ± 0.0 | 10.2 ± 0.11 | 18.2 ± 0.06 | 275 ± 2.2 | 8.45 ± 0.01 | 7.4 ± 0.07 | 2.42 ± 0.06 | 1.28 ± 0.03 | 0.13 ± 0.0 | 0.78 ± 0.05 | 0.06 ± 0.0 | 0.12 ± 0.0 | 0.09 ± 0.0 |
| Sep. 1 | 28.3 ± 0.1 | 16.1 ± 0.2 | 33.1 ± 0.06 | 276 ± 1.4 | 8.4 ± 0.05 | 10.11 ± 0.11 | 2.4 ± 0.06 | 1.43 ± 0.07 | 0.16 ± 0.0 | 0.44 ± 0.02 | 0.08 ± 0.0 | 0.08 ± 0.0 | 0.06 ± 0.0 |
| Sep. 15 | 23.2 ± 0.1 | 7.27 ± 0.06 | 9.3 ± 0.01 | 306 ± 4.2 | 8.4 ± 0.03 | 4.52 ± 0.26 | 2.31 ± 0.11 | 1.77 ± 0.12 | 0.09 ± 0.0 | 0.32 ± 0.01 | 0.05 ± 0.0 | 0.17 ± 0.0 | 0.09 ± 0.0 |
| Sep. 30 | 22.5 ± 0.2 | 7.87 ± 0.06 | 10.2 ± 0.02 | 332 ± 3.8 | 7.9 ± 0.0 | 6.29 ± 0.16 | 2.42 ± 0.11 | 1.9 ± 0.08 | 0.15 ± 0.0 | 0.16 ± 0.0 | 0.03 ± 0.0 | 0.17 ± 0.0 | 0.05 ± 0.0 |
| ANOVA | ** | ** | *** | * | * | *** | * | * | ** | ** | ** | *** | * |
| Sampling date | ACE | Chao1 | Shannon diversity (H') | Simpson diversity (1/D) |
|---|---|---|---|---|
| Jul. 13 | 383 (363,415) | 386 (361,433) | 3.58 (3.55,3.62) | 11 (11,11) |
| Jul. 31 | 409 (386,445) | 405 (380,451) | 4.19 (4.16,4.22) | 25 (25,25) |
| Aug. 16 | 463 (443,495) | 458 (436,498) | 3.99 (3.96,4.03) | 16 (16,16) |
| Sep. 1 | 507 (493,531) | 514 (494,552) | 4.33 (4.31,4.35) | 33 (33,33) |
| Sep. 15 | 459 (436,493) | 454 (429,497) | 4.18 (4.15,4.21) | 25 (25,25) |
| Sep. 30 | 401 (381,435) | 399 (375,442) | 4.43 (4.39,4.46) | 33 (33,33) |
| Sampling date | ACE | Chao 1 | Shannon diversity (H') | Simpson diversity (1/D) |
|---|---|---|---|---|
| Jul. 13 | 233 (219,259) | 238 (219,276) | 3.48 (3.43,3.52) | 12 (14,12) |
| Jul. 31 | 324 (289,374) | 259 (233,307) | 3.51 (3.46,3.56) | 14 (14,14) |
| Aug. 16 | 243 (218,286) | 236 (210,288) | 1.29 (1.26,1.32) | 1.61 (1.61,1.62) |
| Sep. 1 | 327 (309,357) | 322 (303,358) | 4.11 (4.07,4.15) | 25 (25,25) |
| Sep. 15 | 342 (322,374) | 348 (322,396) | 4.04 (4.00,4.09) | 20 (20, 20) |
| Sep. 30 | 256 (228,303) | 247 (218,302) | 2.26 (2.21,2.31) | 3.03 (3.01,3.04) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Jia, J.; Chen, S.; Huang, T.; Wang, Y.; Zhao, Z.; Feng, J.; Hao, H.; Li, S.; Ma, X. Dynamics of Bacterial and Fungal Communities during the Outbreak and Decline of an Algal Bloom in a Drinking Water Reservoir. Int. J. Environ. Res. Public Health 2018, 15, 361. https://doi.org/10.3390/ijerph15020361
Zhang H, Jia J, Chen S, Huang T, Wang Y, Zhao Z, Feng J, Hao H, Li S, Ma X. Dynamics of Bacterial and Fungal Communities during the Outbreak and Decline of an Algal Bloom in a Drinking Water Reservoir. International Journal of Environmental Research and Public Health. 2018; 15(2):361. https://doi.org/10.3390/ijerph15020361
Chicago/Turabian StyleZhang, Haihan, Jingyu Jia, Shengnan Chen, Tinglin Huang, Yue Wang, Zhenfang Zhao, Ji Feng, Huiyan Hao, Sulin Li, and Xinxin Ma. 2018. "Dynamics of Bacterial and Fungal Communities during the Outbreak and Decline of an Algal Bloom in a Drinking Water Reservoir" International Journal of Environmental Research and Public Health 15, no. 2: 361. https://doi.org/10.3390/ijerph15020361
APA StyleZhang, H., Jia, J., Chen, S., Huang, T., Wang, Y., Zhao, Z., Feng, J., Hao, H., Li, S., & Ma, X. (2018). Dynamics of Bacterial and Fungal Communities during the Outbreak and Decline of an Algal Bloom in a Drinking Water Reservoir. International Journal of Environmental Research and Public Health, 15(2), 361. https://doi.org/10.3390/ijerph15020361




