Variation of the Bacterial Community in the Rhizoplane Iron Plaque of the Wetland Plant Typha latifolia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tested Soil and Typha latifolia
2.2. Experimental Design
2.3. Sample Preparation
2.4. DNA Extraction
2.5. Sequencing and Data Analysis
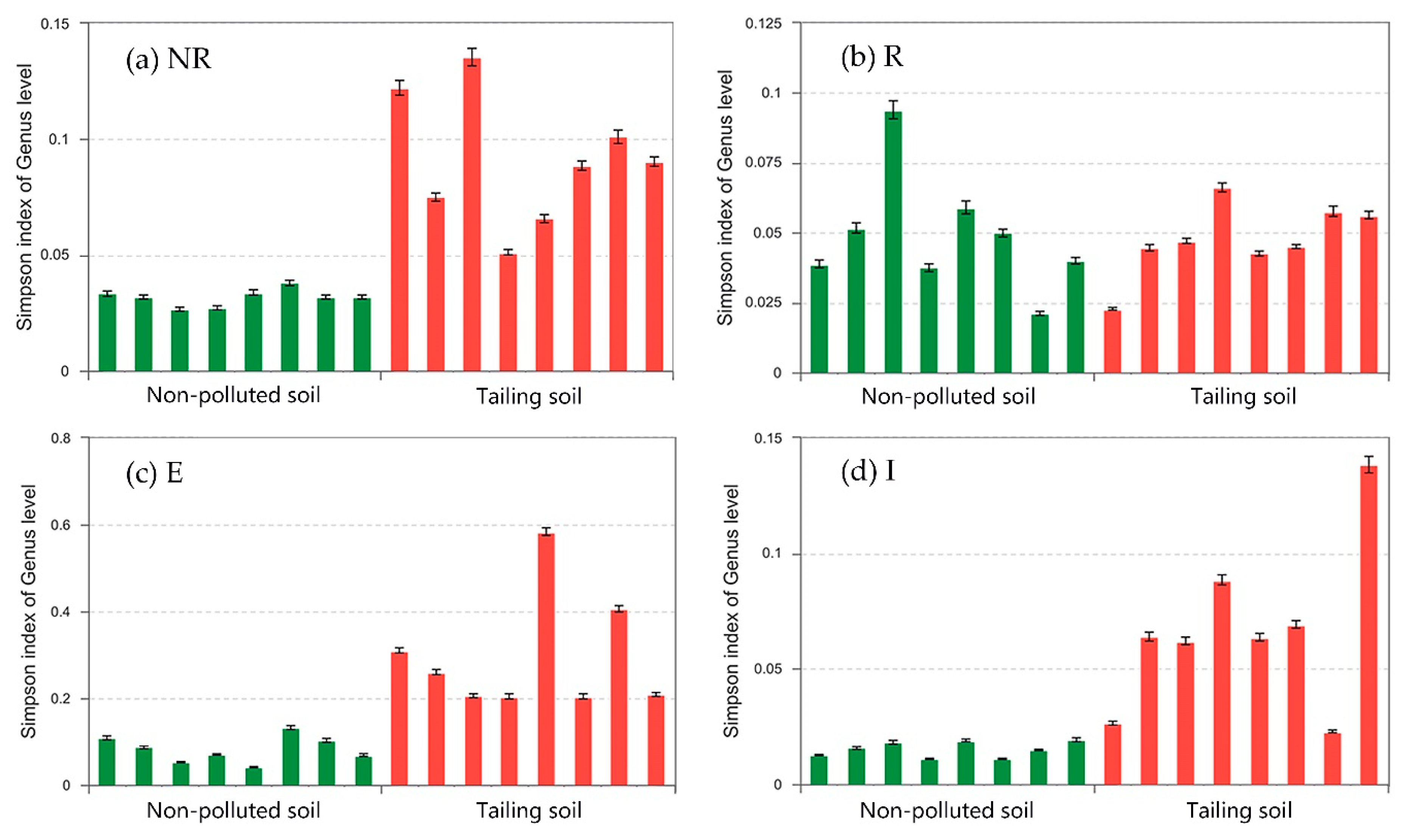
2.6. Simpson Index
2.7. Statistical Methods
3. Results and Discussion
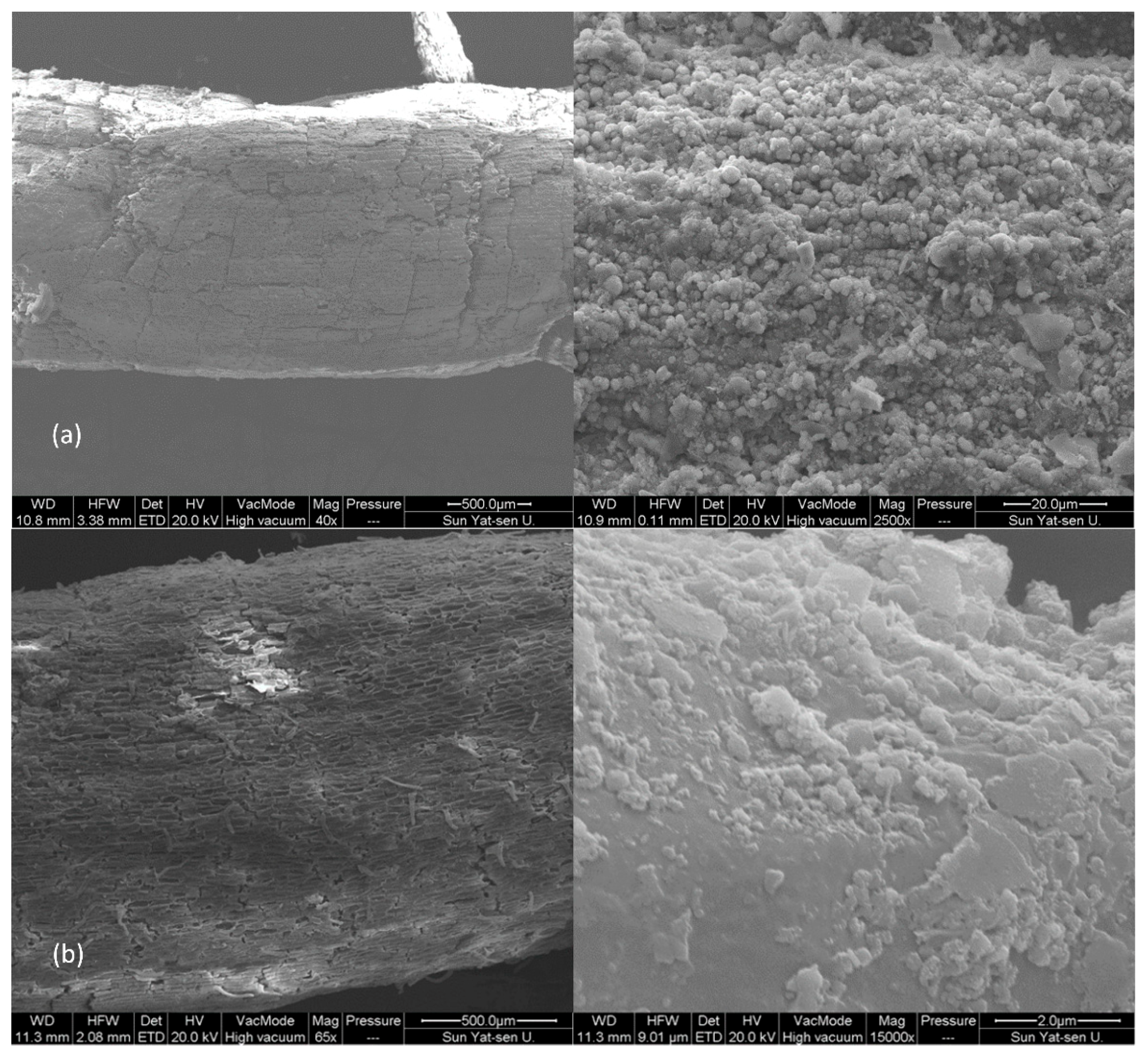
3.1. Separation of Iron Plaque on Roots of Typha latifolia
3.2. Bacterial Community Distribution
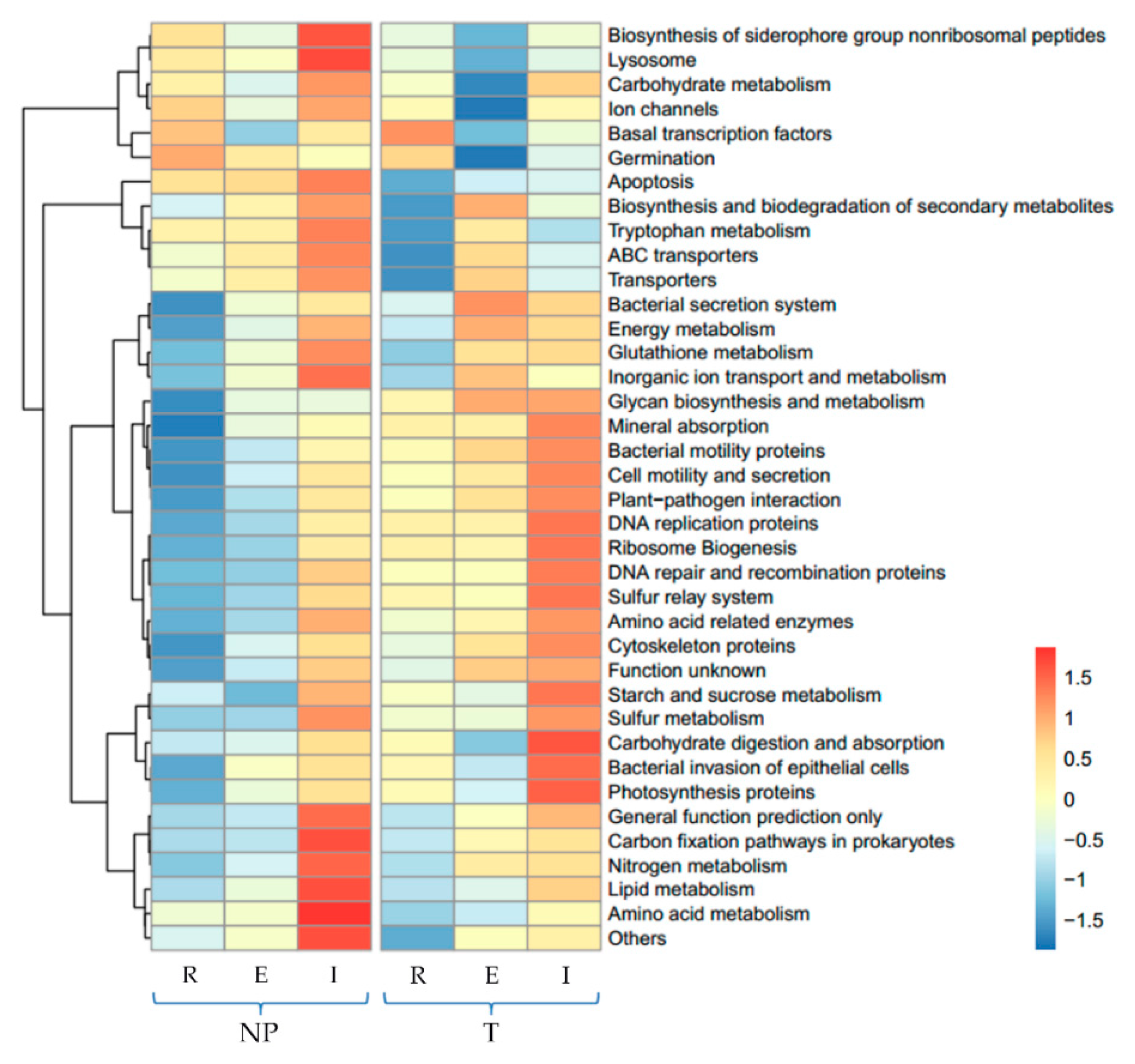
3.3. Predicted Functional Potential of Bacterial Communities
3.4. Distributions of the Dominant Bacterial Phyla
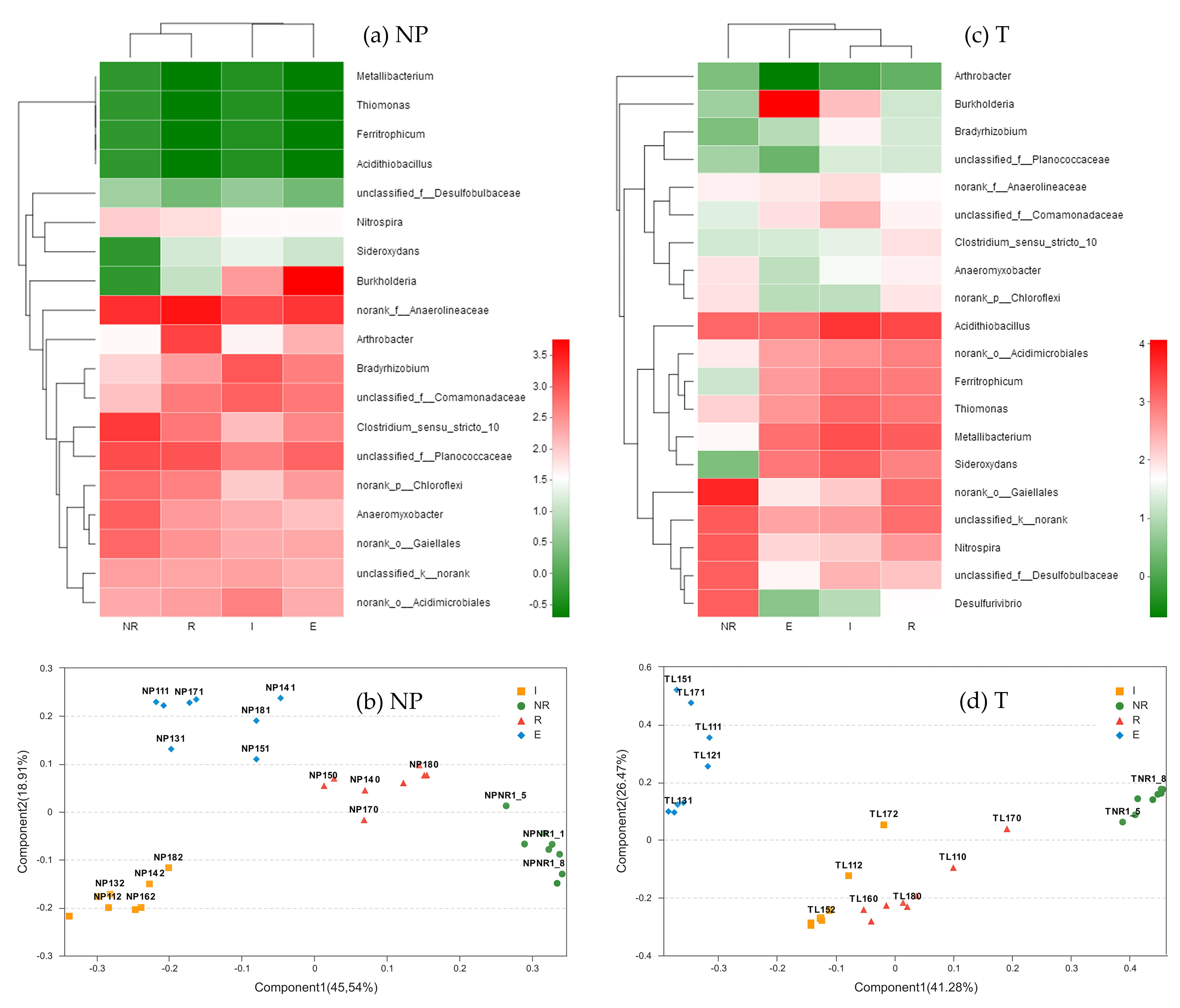
3.5. Dominant Genera of the Bacterial Communities
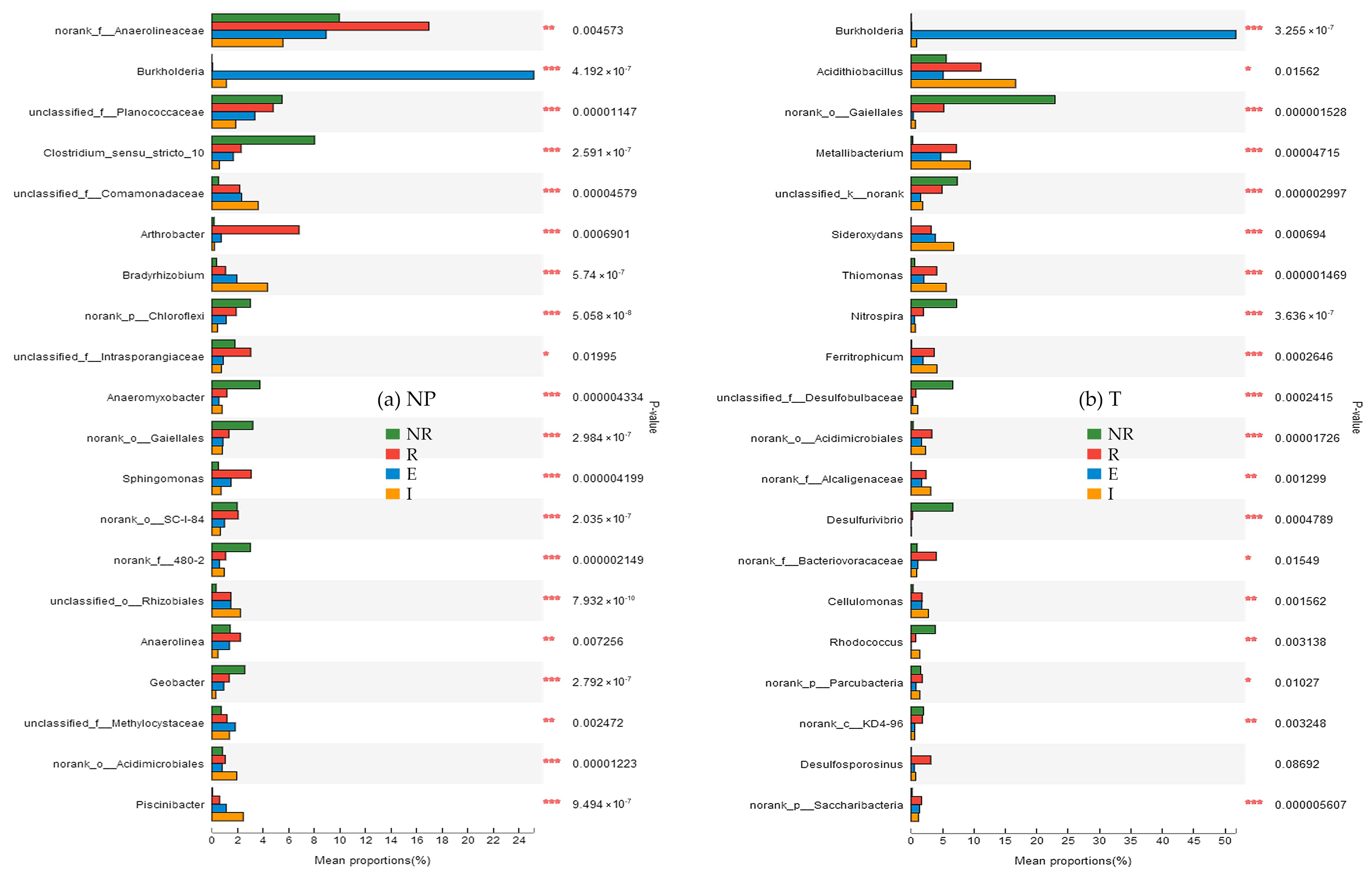
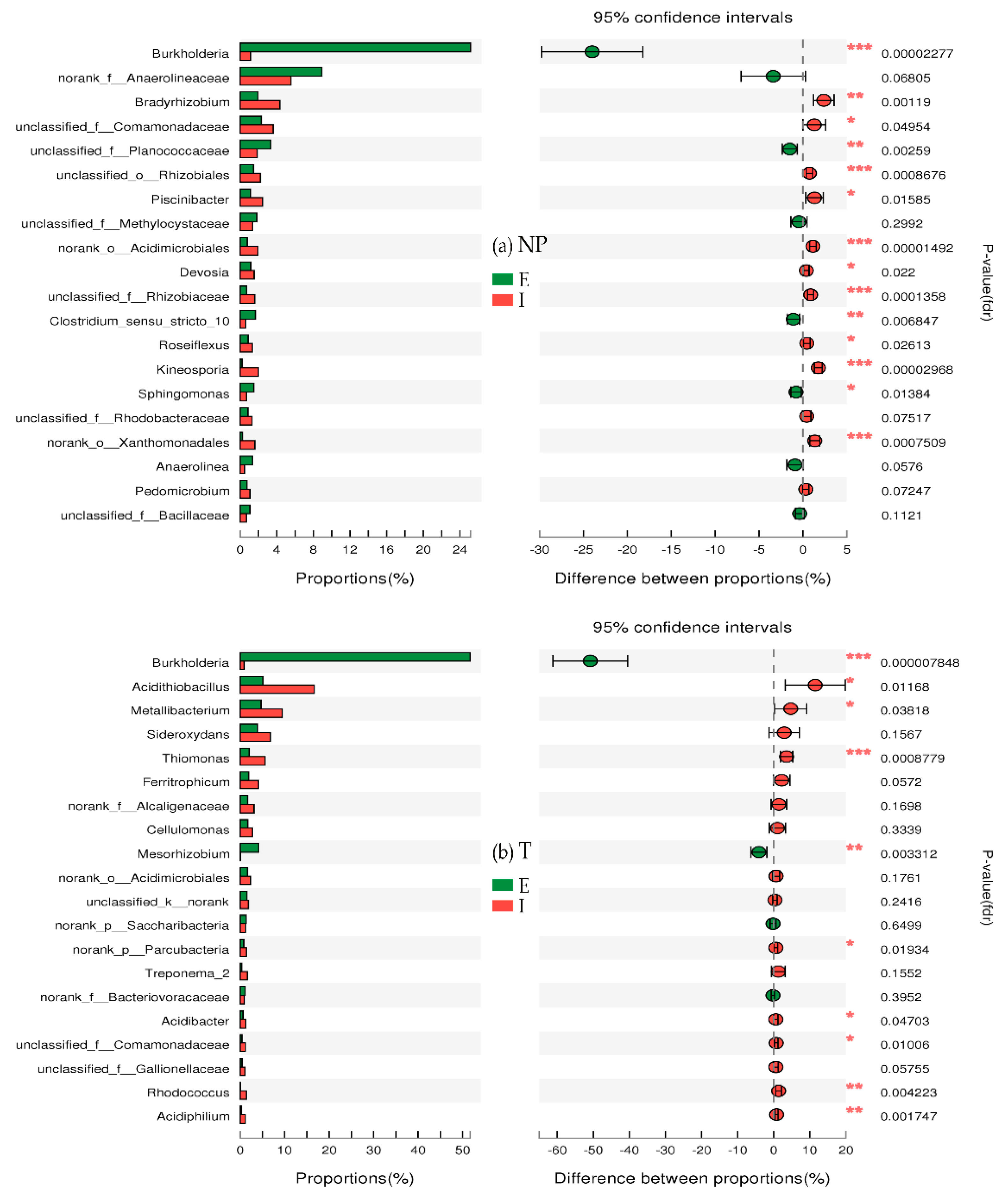
3.6. Genus Differences
3.6.1. Difference in Species Abundance among the NR, R, E, and I
3.6.2. Differences in Species Abundance between E and I
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yan, H.L.; Jing, N.Z.; Zhen, H.Z.; Qiang, Z. Endophytic bacterial diversity in roots of Phragmites australis in constructed Beijing Cuihu Wetland (China). FEMS Microbiol. Lett. 2010, 309, 84–93. [Google Scholar]
- Li, Y.H.; Liu, Q.F.; Liu, Y.; Zhu, J.N.; Zhang, Q. Endophytic bacterial diversity in roots of Typha angustifolia L. In the constructed Beijing Cuihu Wetland (China). Res. Microbiol. 2011, 162, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.F.; Tian, L.I. Pollutants removal and analysis of structure changes of microbial community in integrated vertical-flow constructed wetland. Res. Environ. Sci. 2007, 20, 137–141. [Google Scholar]
- Hansel, C.M.; Fendorf, S.; Sutton, S.; Newville, M. Characterization of fe plaque and associated metals on the roots of mine-waste impacted aquatic plants. Environ. Sci. Technol. 2002, 35, 3863–3868. [Google Scholar] [CrossRef]
- Stubner, S.; Wind, T.; Conrad, R. Sulfur oxidation in rice field soil: Activity, enumeration, isolation andcharacterization of thiosulfate-oxidizing bacteria. Syst. Appl. Microbiol. 1998, 21, 569–578. [Google Scholar] [CrossRef]
- Emerson, D.; Weiss, J.V.; Megonigal, J.P. Iron-oxidizing bacteria are associated with ferric hydroxide precipitates (Fe-plaque) on the roots of wetland plants. Appl. Environ. Microbiol. 1999, 65, 2758–2761. [Google Scholar] [PubMed]
- Wind, T.; Stubner, S.; Conrad, R. Sulfate-reducing bacteria in rice field soil and on rice roots. Syst. Appl. Microbiol. 1999, 22, 269. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.Y.; Chen, X.; Luo, A.C. Investigation on the microbial functional groups characteristics of rhizoshphere of wetland plants in mimic wastewater treatment. J. Agro-Environ. Sci. 2010, 29, 764–768. [Google Scholar]
- Zhang, X.; Chen, L.; Liu, X.; Wang, C.; Chen, X.; Xu, G.; Deng, K. Synergic degradation of diesel by scirpus triqueter and its endophytic bacteria. Environ. Sci. Pollut. Res. 2014, 21, 8198–8205. [Google Scholar] [CrossRef] [PubMed]
- He, L.Y.; Chen, Z.J.; Ren, G.D.; Zhang, Y.F.; Meng, Q.; Sheng, X.F. Increased cadmium and lead uptake of a cadmium hyperaccumulator tomato by cadmium-resistant bacteria. Ecotoxicol. Env. Saf. 2009, 72, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Sasmaz, A.; Obek, E.; Hasar, H. The accumulation of heavy metals in typha latifolia l. Grown in a stream carrying secondary effluent. Ecol. Eng. 2008, 33, 278–284. [Google Scholar] [CrossRef]
- Deng, H.; Zhi-Hong, Y.E.; Huang, M.H. Characteristics of radial oxygen loss from root of wetland plants. J. East China Norm. Univ. 2007, 30, 69–76. [Google Scholar]
- Cheng, X.Y.; Wang, M.; Zhang, C.F.; Wang, S.Q.; Chen, Z.H. Relationships between plant photosynthesis, radial oxygen loss and nutrient removal in constructed wetland microcosms. Biochem. Syst. Ecol. 2014, 54, 299–306. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver, L.V.T.E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; Mcdonald, D.; Dan, K.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, E.H. The interpretation of interaction in contingency tables. J. R. Stat. Soc. 1951, 13, 238–241. [Google Scholar]
- Morris, C.E.; Monier, J.M. The ecological significance of biofilm formation by plant-associated bacteria. Annu. Rev. Phytopathol. 2003, 41, 429–453. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.J.; Crowder, A.A. Uptake and accumulation of heavy metals by typha latifolia in wetlands. Can. J. Bot. 1983, 61, 63–73. [Google Scholar] [CrossRef]
- Chabbi, A.; Hines, M.E.; Rumpel, C. The role of organic carbon excretion by bulbous rush roots and its turnover and utilization by bacteria under iron plaques in extremely acid sediments. Environ. Exp. Bot. 2001, 46, 237–245. [Google Scholar] [CrossRef]
- Hou, D.; Wang, K.; Liu, T.; Wang, H.; Lin, Z.; Qian, J.; Lu, L.; Tian, S. Unique rhizosphere micro-characteristics facilitate phytoextraction of multiple metals in soil by the hyperaccumulating plant Sedum alfredii. Environ. Sci. Technol. 2017, 51, 5675. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Ding, Q.B.; Chao, Y.Q.; Wei, X.G.; Wang, S.Z.; Qiu, R.L. Structural development and assembly patterns of the root-associated microbiomes during phytoremediation. Sci. Total Environ. 2018, 644, 1591–1601. [Google Scholar] [CrossRef]
- Sibanda, T.; Selvarajan, R.; Msagati, T.; Venkatachalam, S.; Meddows-Taylor, S. Defunct gold mine tailings are natural reservoir for unique bacterial communities revealed by high-throughput sequencing analysis. Sci. Total Environ. 2018, 650, 2199–2209. [Google Scholar] [CrossRef] [PubMed]
- Quaiser, A.; Ochsenreiter, T.; Lanz, C.; Schuster, S.C.; Treusch, A.H.; Eck, J.; Schleper, C. Acidobacteria form a coherent but highly diverse group within the bacterial domain: Evidence from environmental genomics. Mol. Microbiol. 2003, 50, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Barns, S.M.; Cain, E.C.; Sommerville, L.; Kuske, C.R. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 2007, 73, 3113–3116. [Google Scholar] [CrossRef] [PubMed]
- Nelson, W.C.; Stegen, J.C. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 2015, 6, 713. [Google Scholar] [CrossRef] [PubMed]
- Castelle, C.J.; Brown, C.T.; Thomas, B.C.; Williams, K.H.; Banfield, J.F. Unusual respiratory capacity and nitrogen metabolism in a Parcubacterium (OD1) of the candidate phyla radiation. Sci. Rep. 2017, 7, 40101. [Google Scholar] [CrossRef] [PubMed]
- Dourado, M.N.; Martins, P.F.; Quecine, M.C.; Piotto, F.A.; Souza, L.A.; Franco, M.R.; Tezotto, T.; Azevedo, R.A. Burkholderia sp. SCMS54 reduces cadmium toxicity and promotes growth in tomato. Ann. Appl. Biol. 2013, 163, 494–507. [Google Scholar]
- Jiang, C.Y.; Sheng, X.F.; Qian, M.; Wang, Q.Y. Isolation and characterization of a heavy metal-resistant Burkholderia sp. From heavy metal-contaminated paddy field soil and its potential in promoting plant growth and heavy metal accumulation in metal-polluted soil. Chemosphere 2008, 72, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Xiao, T.; Sun, M.; Dong, Y.; Ning, Z.; Xiao, E.; Tang, S.; Li, J. Diversity of the sediment microbial community in the Aha watershed (Southwest China) in response to acid mine drainage pollution gradients. Appl. Environ. Microbiol. 2015, 81, 4874–4884. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Xiao, E.; Krumins, V.; Dong, Y.; Xiao, T.; Ning, Z.; Chen, H.; Xiao, Q. Characterization of the microbial community composition and the distribution of Fe-metabolizing bacteria in a creek contaminated by acid mine drainage. Appl. Microbiol. Biotechnol. 2016, 100, 1–13. [Google Scholar] [CrossRef] [PubMed]
- ArsãNe-Ploetze, F.; Koechler, S.; Marchal, M.; Coppã©E, J.Y.; Chandler, M.; Bonnefoy, V.; Brochier-Armanet, C.; Barakat, M.; Barbe, V.; Battaglia-Brunet, F. Structure, function, and evolution of the Thiomonas spp. Genome. PLoS Genet. 2010, 6, e1000859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedrich, C.; Bardischewsky, F.; Rother, D.A.; Fischer, J. Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 2005, 8, 253. [Google Scholar] [CrossRef] [PubMed]
- Frigaard, N.U.; Dahl, C. Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 2009, 54, 103. [Google Scholar] [PubMed]
- Ziegler, S.; Waidner, B.; Itoh, T.; Schumann, P.; Spring, S.; Gescher, J. Metallibacterium scheffleri gen. nov., sp. nov., an alkalinizing gammaproteobacterium isolated from an acidic biofilm. Int. J. Syst. Evol. Microbiol. 2013, 63, 1499. [Google Scholar] [CrossRef] [PubMed]
- Jie, H.U.; Xiaohong, H.E.; Daping amp, L.I.; Qiang, L. Progress in research of Sphingomonas. Chin. J. Appl. Environ. Boil. 2007, 13, 431–437. [Google Scholar]
- White, D.C.; Sutton, S.D.; Ringelberg, D.B. The genus Sphingomonas: Physiology and ecology. Curr. Opin. Biotechnol. 1996, 7, 301. [Google Scholar] [CrossRef]
- Zhao, H.; Zhao, J.; Li, F.; Li, X. Performance of denitrifying microbial fuel cell with biocathode over nitrite. Front. Microbiol. 2016, 7, 344. [Google Scholar] [CrossRef] [PubMed]
- Bautista, V.V.; Monsalud, R.G.; Yokota, A. Devosia yakushimensis sp. nov., isolated from root nodules of Pueraria lobata (Willd.) Ohwi. Int. J. Syst. Evol. Microbiol. 2010, 60, 627. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bond, P.L.; Van Nostrand, J.D.; Zhou, J.; Huang, L. From lithotroph- to organotroph-dominant: Directional shift of microbial community in sulphidic tailings during phytostabilization. Sci. Rep. 2015, 5, 12978. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.; Liu, W.; Chen, Y.; Chen, W.; Zhao, L.; Ding, Q.; Wang, S.; Tang, Y.T.; Zhang, T.; Qiu, R.L. Structure, variation, and co-occurrence of soil microbial communities in abandoned sites of a rare earth elements mine. Environ. Sci. Technol. 2016, 50, 11481. [Google Scholar] [CrossRef] [PubMed]
- Van, D.M.; Marcel, T.J.; Klatt, C.G.; Wood, J.; Bryant, D.A.; Bateson, M.M.; Lammerts, L.; Schouten, S.; Damste, J.S.S.; Madigan, M.T.; et al. Cultivation and genomic, nutritional, and lipid biomarker characterization of roseiflexus strains closely related to predominant in situ populations inhabiting yellowstone hot spring microbial mats. J. Bacteriol. 2010, 192, 3033. [Google Scholar]
- Hao, X.; Xie, P.; Zhu, Y.G.; Taghavi, S.; Wei, G.; Rensing, C. Copper tolerance mechanisms of mesorhizobium amorphae and its role in aiding phytostabilization by robinia pseudoacacia in copper contaminated soil. Environ. Sci. Technol. 2015, 49, 2328–2340. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Zhou, Y.; Fu, S.; Yang, Z.; Zhang, C.; Cai, L. Research on the comprehensive utilization of dabaoshan polymetallic sulfide tailings. Met. Mine. 2009, 7, 164–168. [Google Scholar]
- Harrison, A.P. Acidiphilium cryptum gen. nov., sp. nov., heterotrophic bacterium from acidic mineral environments. Int. J. Syst. Bacteriol. 1981, 31, 327–332. [Google Scholar] [CrossRef]
- Toni, A.M.; Bridge, D.B.J. Reductive dissolution of ferric iron minerals by Acidiphilium SJH. Geomicrobiol. J. 2000, 17, 193–206. [Google Scholar]


| Physicochemical Property | Soil | ||
|---|---|---|---|
| T (Tailing Soil) | NP (Non-Polluted Soil) | ||
| pH | 3.16 ± 0.02 | 5.96 ± 0.06 | |
| Total (mg/kg) | C | 500.00 ± 100.00 | 12,766.67 ± 650.64 |
| N | n.d. | 1066.67 ± 57.74 | |
| S | 101,200.00 ± 2179.45 | n.d. | |
| Total (mg/kg) | Pb | 78.48 ± 5.91 | 40.75 ± 2.13 |
| Zn | 492.05 ± 31.88 | 76.01 ± 6.02 | |
| Cu | 3048.95 ± 153.24 | 32.40 ± 0.76 | |
| Cd | 3.26 ± 0.37 | 0.37 ± 0.09 | |
| Fe | 15,851.22 ± 2443.78 | 21,094.82 ± 280.14 | |
| NH4NO3-extractable (mg/kg) | Pb | 2.26 ± 0.42 | 0.02 ± 0.02 |
| Zn | 96.23 ± 4.06 | 0.27 ± 0.13 | |
| Cu | 99.67 ± 3.89 | 0.29 ± 1.60 | |
| Cd | 0.33 ± 0.02 | 0.03 ± 0.00 | |
| Fe | 835.69 ± 17.79 | 0.63 ± 0.26 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chi, H.; Yang, L.; Yang, W.; Li, Y.; Chen, Z.; Huang, L.; Chao, Y.; Qiu, R.; Wang, S. Variation of the Bacterial Community in the Rhizoplane Iron Plaque of the Wetland Plant Typha latifolia. Int. J. Environ. Res. Public Health 2018, 15, 2610. https://doi.org/10.3390/ijerph15122610
Chi H, Yang L, Yang W, Li Y, Chen Z, Huang L, Chao Y, Qiu R, Wang S. Variation of the Bacterial Community in the Rhizoplane Iron Plaque of the Wetland Plant Typha latifolia. International Journal of Environmental Research and Public Health. 2018; 15(12):2610. https://doi.org/10.3390/ijerph15122610
Chicago/Turabian StyleChi, Haochun, Lu Yang, Wenjing Yang, Yuanyuan Li, Ziwu Chen, Lige Huang, Yuanqing Chao, Rongliang Qiu, and Shizhong Wang. 2018. "Variation of the Bacterial Community in the Rhizoplane Iron Plaque of the Wetland Plant Typha latifolia" International Journal of Environmental Research and Public Health 15, no. 12: 2610. https://doi.org/10.3390/ijerph15122610
APA StyleChi, H., Yang, L., Yang, W., Li, Y., Chen, Z., Huang, L., Chao, Y., Qiu, R., & Wang, S. (2018). Variation of the Bacterial Community in the Rhizoplane Iron Plaque of the Wetland Plant Typha latifolia. International Journal of Environmental Research and Public Health, 15(12), 2610. https://doi.org/10.3390/ijerph15122610





