Multidrug-Resistant Bacteria Isolated from Surface Water in Bassaseachic Falls National Park, Mexico
Abstract
:1. Introduction
2. Experimental Section
2.1. Sample Collection
2.2. Microbiological Analysis
2.2.1. Total/Fecal Coliform Determination
2.2.2. Microbial Isolation
2.3. Biochemical Identification
2.4. Antibiotic Resistance Analyses
2.5. Statistical Analysis
3. Results
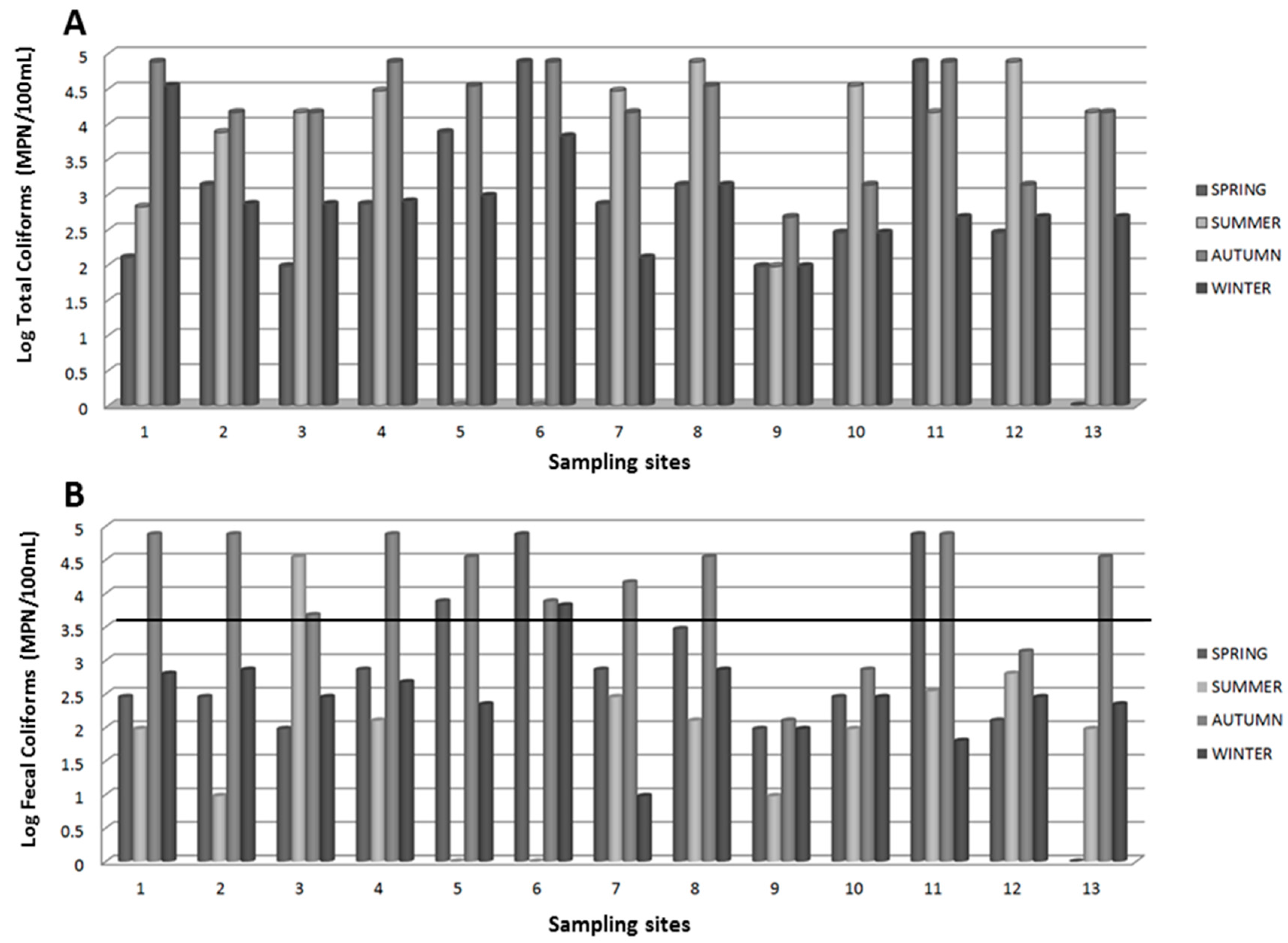
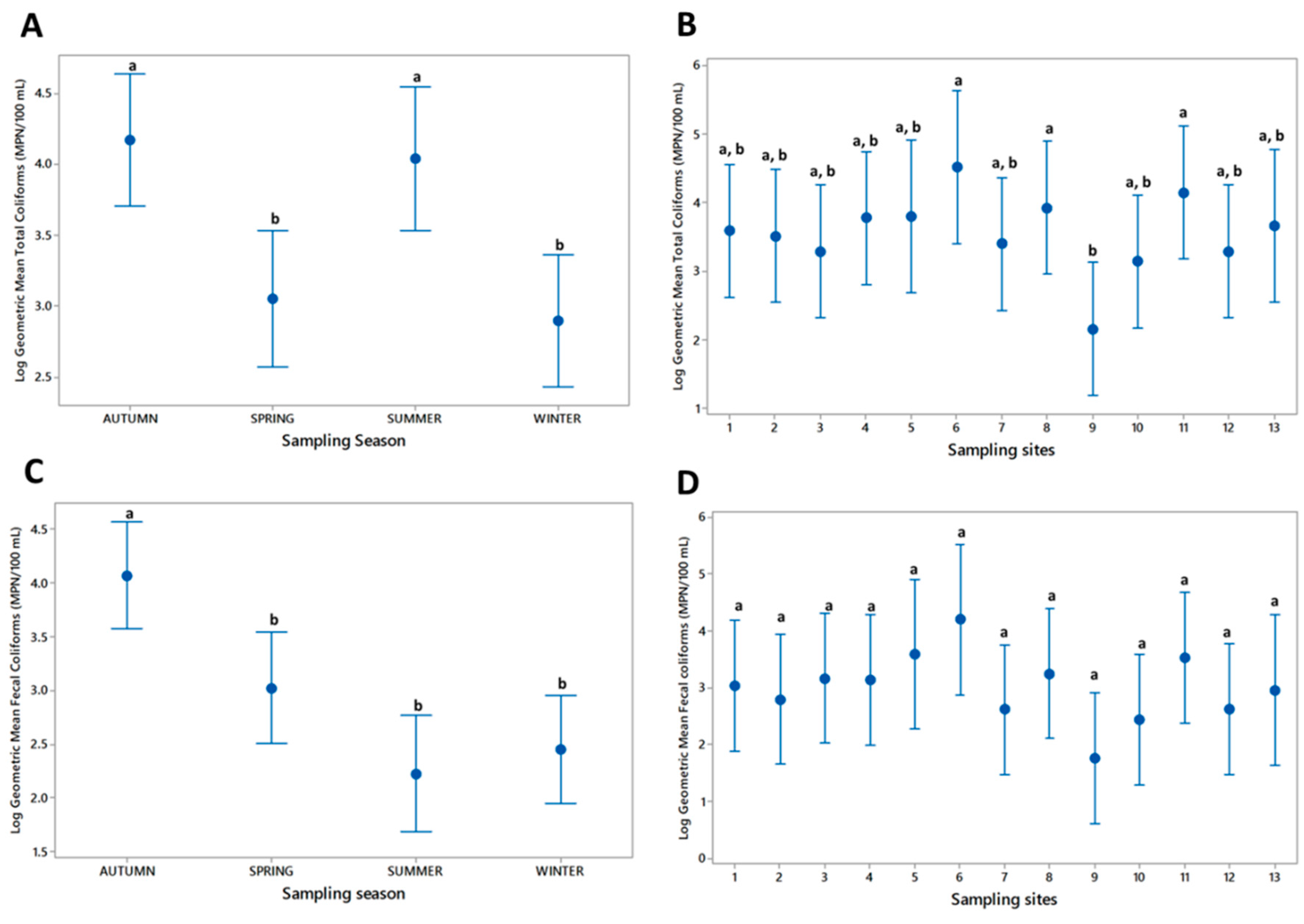
3.1. Microbiological Analysis
3.2. Biochemical and Antibiotic Resistance Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Baquero, F.; Martínez, J.L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Korzeniewska, E.; Korzeniewska, A.; Harnisz, M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013, 91, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Figueras, M.J.; Borrego, J.J. New perspectives in monitoring drinking water microbial quality. Int. J. Environ. Res. Public Health 2010, 7, 4179–4202. [Google Scholar] [CrossRef] [PubMed]
- Ashbolt, N.J. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology 2004, 198, 229–238. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Diarrhoeal Disease Fact Sheet. Available online: http://www.who.int/mediacentre/factsheets/fs330/en/ (accessed on 31 March 2016).
- Amdiouni, H.; Maunula, L.; Hajjami, K.; Faouzi, A.; Soukri, A.; Nourlil, J. Recovery comparison of two virus concentration methods from wastewater using cell culture and real-time PCR. Curr. Microbiol. 2012, 65, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Edqvist, L.; Pedersen, K. Antimicrobials as growth promoters: Resistance to common sense. In Late Lessons from Early Warnings: The Precautionaryprinciple 1896–2000; Harremoës, P., Gee, D., MacGarvin, M., Stirling, A., Keys, J., Wynne, B., Guedes Vaz, S., Eds.; European Environmental Agency: Copenhagen, Denmark, 2001; pp. 93–100. [Google Scholar]
- Brown, M.G.; Balkwill, D.L. Antibiotic resistance in bacteria isolated from the deep terrestrial subsurface. Microb. Ecol. 2009, 57, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Gesche, E.; Vallejos, A.; Sáez, M. Eficiencia de anaerobios sulfito-reductores como indicadores de calidad sanitaria de agua. Método de Número Más Probable (NMP). Arch. Med. Vet. 2003, 35, 99–107. [Google Scholar] [CrossRef]
- Bush, K.; Courvalin, P.; Dantas, G.; Davies, J.; Eisenstein, B.; Huovinen, P.; Jacoby, G.A.; Kishony, R.; Kreiswirth, B.N.; Kutter, E.; et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011, 9, 894–896. [Google Scholar] [CrossRef] [PubMed]
- Port, J.A.; Cullen, A.C.; Wallace, J.C.; Smith, M.N.; Faustman, E.M. Metagenomic frameworks for monitoring antibiotic resistance in aquatic environments. Environ. Health Perspect. 2014, 122, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Diario Oficial de la Federación (1994). NOM-014-SSA1-1993. Procedimientos Sanitarios para el Muestreo de Agua para uso y Consumo Humano en Sistemas de Abastecimiento de Agua Públicos y Privados. Secretaría de Salud. México, D.F. Available online: http://www.salud.gob.mx/unidades/cdi/nom/127ssa14.html (accessed on 14 June 2016).
- Diario Oficial de la Federación (2005). Proyecto PROY-NMX-AA-042-SCFI-2005. Calidad del Agua.-Determinación del Número más Probable (NMP) de Coliformes Totales, Coliformes Fecales (Termotolerantes) y Escherichia coli Presuntiva (Cancelará a la NMX-AA-042-1987). México, D.F. Available online: http://legismex.mty.itesm.mx/normas/ aa/nmx-AA-05/proy-nmx-aa-042-scfi-2005.pdf (accessed on 14 June 2016).
- Al-Bayatti, K.K.; Al-Arajy, K.H.; Al-Nuaemy, S.H. Bacteriological and physicochemical studies on Tigris River near the water purification stations within Baghdad Province. J. Environ. Public Health 2012, 2012, 695253. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.B.; Smith, H.L.; Ogg, J.E. Isolation of non-O1 Vibrio cholerae serovars from surface waters in western Colorado. Appl. Environ. Microbiol. 1986, 51, 1216–1219. [Google Scholar] [PubMed]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved Standard. CLSI document M07-A10; Wayne, PA, USA, 2015, 10th ed. Available online: http://shop.clsi.org/site/Sample_pdf/M07A10_sample.pdf (accessed on 31 March 2016).
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Document M100-S24; Wayne, PA, USA, 2014. Available online: http://ncipd.org/control/images/NCIPD_docs/CLSI_M100-S24.pdf (accessed on 31 March 2016).
- Diario Oficial de la Federación. (1997) DOF-001-ECOL-1996, que Establece los Límites Máximos Permisibles de Contaminantes en las Descargas de Aguas Residuales a los Sistemas en Aguas y Bienes Nacionales. Gobierno Constitucional de los Estados Unidos Mexicanos. México D.F. Available online: http://dof.gob.mx/nota_detalle.php?codigo=4863829&fecha=06/01/1997 (accessed on 14 June 2016).
- Agger, W.A.; McCormick, J.D.; Gurwith, M.J. Clinical and microbiological features of Aeromonas hydrophila-associated diarrhea. J. Clin. Microbiol. 1985, 21, 909–913. [Google Scholar] [PubMed]
- Cabral, J.P. Water microbiology. Bacterial pathogens and water. Int. J. Environ. Res. Public Health 2010, 7, 3657–3703. [Google Scholar] [CrossRef] [PubMed]
- Abbott, S.L.; Janda, J.M. Revisiting bacterial gastroenteritis: Issues, possible approaches, and an ever-expanding list of etiologic agents, Part II. Clin. Microbiol. Newslett. 2011, 33, 79–86. [Google Scholar] [CrossRef]
- Threlfall, E.J. Antimicrobial drug resistance in Salmonella: Problems and perspectives in food- and water-borne infections. FEMS Microbiol. Rev. 2002, 26, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbot, S.L. Uncommon enterobacterial genera associated with clinical specimens. In The Enterobacteria, 2nd ed.; American Society Microbiology: Washington, DC, USA, 2006; pp. 357–375. [Google Scholar]
- Thompson, J.S.; Gravel, M.J. Family outbreak of gastroenteritis due to Yersinia enterocolitica serotype 0:3 from well water. Can. J. Microbiol. 1986, 32, 700–701. [Google Scholar] [CrossRef] [PubMed]
- Poblete, J. Pasteurella multocida bacteremia: Concern remains even in the absence of direct animal contact. Inf. Dis. Clin. Pract. 2009, 17, 77. [Google Scholar] [CrossRef]
- Heéger, Z.; Vargha, M.; Márialigeti, K. Detection of potentially pathogenic bacteria in the drinking water distribution system of a hospital in Hungary. Clin. Microbiol. Infect. 2010, 16, 89–92. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Davino, A.M.; Melo, M.B.; Caffaro Filho, R.A. Assessing the sources of high fecal coliform levels at an urban tropical beach. Braz. J. Microbiol. 2015, 46, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.E.; Mansour, A.; Shaheen, H.; Amine, M.; Riddle, M.S.; Young, S.Y.; Sebeny, P.; Levin, S. The impact of household hygiene on the risk of bacterial diarrhea among Egyptian children in rural areas, 2004–2007. J. Infect. Dev. Countr. 2014, 8, 1541–1551. [Google Scholar] [CrossRef] [PubMed]
- Payment, P.; Locas, A. Pathogens in water: Value and limits of correlation with microbial indicators. Ground Water 2011, 49, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Van den Bogaard, A.E.; Stobberingh, E.E. Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicrob. Agents 2000, 14, 327–335. [Google Scholar] [CrossRef]
- Girones, R.; Ferrús, M.A.; Alonso, J.L.; Rodriguez-Manzano, J.; Calgua, B.; de Abreu Corrêa, A.; Hundesa, A.; Carratala, A.; Bofill-Mas, S. Molecular detection of pathogens in water—The pros and cons of molecular techniques. Water Res. 2010, 44, 4325–4339. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, J.P.; Toze, S.G. Human pathogens and their indicators in biosolids: A literature review. Environ. Int. 2009, 35, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Grant, A.; Hashem, F.; Parveen, S. Salmonella and Campylobacter: Antimicrobial resistance and bacteriophage control in poultry. Food Microbiol. 2016, 53, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Titilawo, Y.; Obi, L.; Okoh, A. Occurrence of virulence gene signatures associated with diarrhoeagenic and non-diarrhoeagenic pathovars of Escherichia coli isolates from some selected rivers in South-Western Nigeria. BMC Microbiol. 2015, 15, 204. [Google Scholar] [CrossRef] [PubMed]
- Amit, A.; Kumar, A.; Kumar, M.; Rahal, A. Multidrug resistant pathogenic Escherichia coli status in water sources and Yamuna River in and around Mathura, India. Pak. J. Biol. Sci. 2014, 17, 540–544. [Google Scholar] [CrossRef]
- Miranda, C.D.; Zemelman, R. Antimicrobial multiresistance in bacteria isolated from freshwater Chilean salmon farms. Sci. Total Environ. 2002, 293, 207–218. [Google Scholar] [CrossRef]
- Podschun, R.; Pietsch, S.; Höller, C.; Ullmann, U. Incidence of klebsiella species in surface waters and their expression of virulence factors. Appl. Environ. Microbiol. 2001, 67, 3325–3327. [Google Scholar] [CrossRef] [PubMed]
- Sayah, R.S.; Kaneene, J.B.; Johnson, Y.; Miller, R. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 2005, 71, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Debarry, J.; Garn, H.; Hanuszkiewicz, A.; Dickgreber, N.; Blümer, N.; von Mutius, E.; Bufe, A.; Gatermann, S.; Renz, H.; Holst, O.; et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J. Allergy Clin. Immunol. 2007, 119, 1514–1521. [Google Scholar] [CrossRef] [PubMed]
- Hollis, D.G.; Hickman, F.W.; Fanning, G.R.; Farmer, J.J., III; Weaver, R.E.; Brenner, D.J. Tatumella ptyseos gen. nov., sp. nov., A member of the family Enterobacteriaceae found in clinical specimens. J. Clin. Microbiol. 1981, 14, 79–88. [Google Scholar] [PubMed]
- Toranzos, G.; Marcos, R. Human enteric pathogens and soil borne-disease. In Soil Biochemistry; Bollag, J.-M., Stotzky, G., Eds.; Marcel Dekker: New York, NY, USA, 2000; pp. 461–481. [Google Scholar]
- Kellfy, B.G.; Vespermann, A.; Bolton, D.J. Gene transfer events and their occurrence in selected environments. Food Chem. Toxicol. 2009, 47, 978–983. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.W.; Olivares-Rieumont, S.; Knapp, C.W.; Lima, L.; Werner, D.; Bowen, E. Antibiotic resistance gene abundances associated with waste discharges to the Almendares River near Havana, Cuba. Environ. Sci. Technol. 2011, 45, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Larsson, D.G. Antibiotics in the environment. Upsala J. Med. Sci. 2014, 119, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.; Marhova, M.; Gochev, V.; Tsankova, M.; Trifonova, S. Antibiotic resistance of Gram-negative benthic bacteria isolated from the sediments of Kardzhali Dam (Bulgaria). Biotechnol. Biotechnol. Equip. 2015, 29, 274–280. [Google Scholar] [CrossRef] [PubMed]
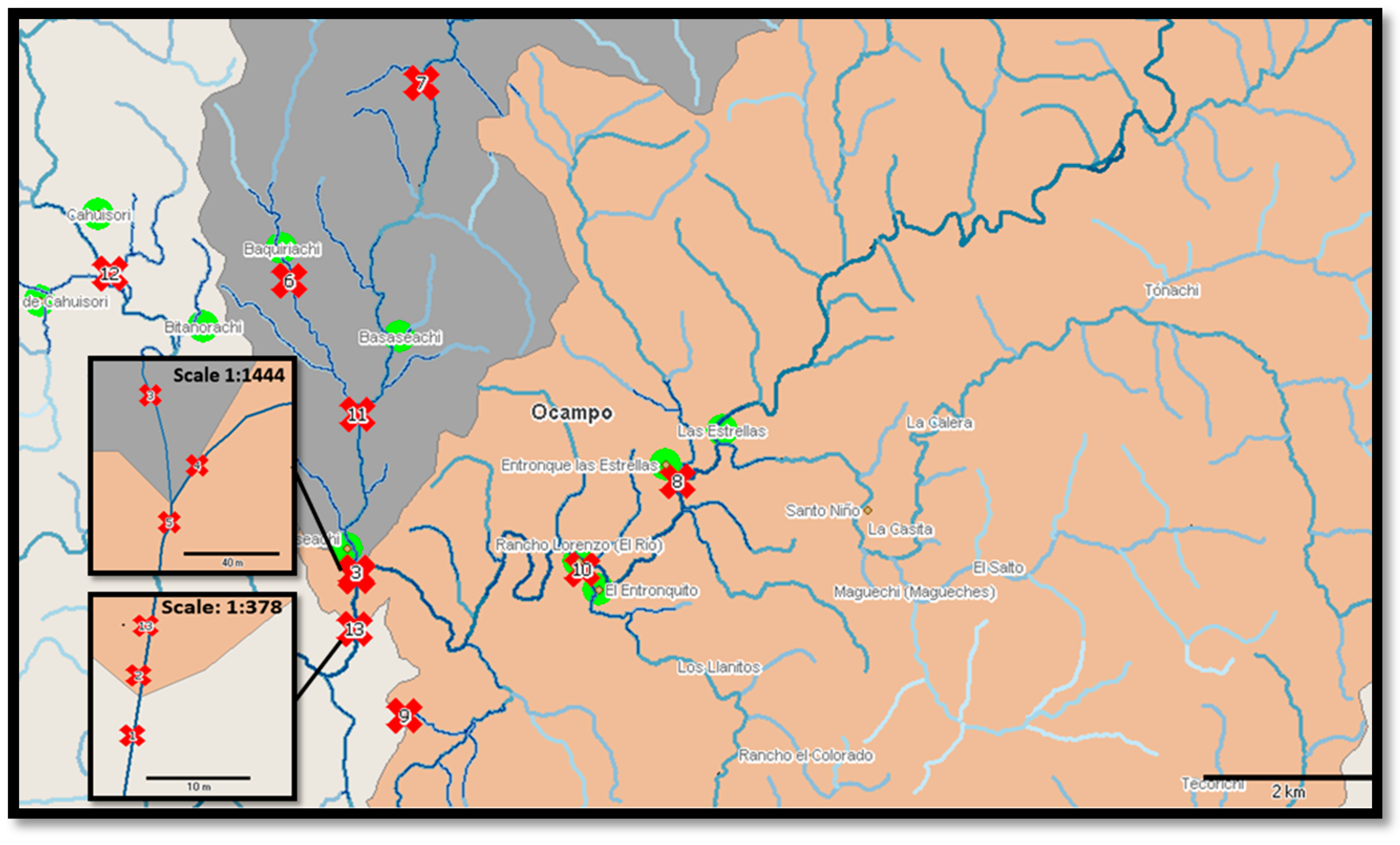

| Sample Points | Geographical Coordinates | ||
|---|---|---|---|
| Sample | Sample Site | North (N) | West (W) |
| 1 | Basaseachi waterfall well | 28°10′26.95″N | 108°12′44.99″W |
| 2 | “La ventana” waterfall | 28°10′27.17″N | 108°12′44.88″W |
| 3 | “El Durazno” river | 28°10′48.24″N | 108°12′45.09″W |
| 4 | Basaseachi river | 28°10′49.19″N | 108°12′45.86″W |
| 5 | Y. Basaseachi junction of the river and Durazno river. | 28°10′47.45″N | 108°12′45.49″W |
| 6 | “Baquiriachi” stream | 28°12′40.80″N | 108°13′20.49″W |
| 7 | Basaseachi water supply stream | 28°13′59.55″N | 108°12′26.07″W |
| 8 | “Las Estrellas” stream | 28°11′30.06″N | 108°10′27.09″W |
| 9 | Visitors center of Bassaseachic Falls National Park | 28°09′54.7″N | 108°12′21.7″W |
| 10 | “Betorachi” stream | 28°10′54.08″N | 108°11′6.84″W |
| 11 | Creek next to the oxidation pond | 28°11′50.25″N | 108°12′47.88″W |
| 12 | “Cahuisori” stream | 28°12′40.58″N | 108°14′38.92″W |
| 13 | Belvedere of Basaseachi waterfall | 28°10′27.36″N | 108°12′44.97″W |
| Family | Isolate Identification and References 1 | Number of Isolates | Sampling Site | Sampling Season |
|---|---|---|---|---|
| Enterobacteriaceae | Aeromonas hydrophila (Chester) Stanier [20] | 1 | 3 | Autumn |
| Citrobacter freundii Werkman & Gillen [21,22] | 2 | 2,5 | Summer–Winter | |
| Enterobacter cloacae Jordan/Hormaeche [21] | 3 | 1,3,6 | Autumn–Winter | |
| Escherichia coli Escherich [21] | 7 | 1,6,11,13 | Spring-Summer-Autumn-Winter | |
| Hafnia alvei Møller [22] | 3 | 1,2,3 | Summer-Autumn | |
| Klebsiella oxytoca Flügge/Lautrop [21] | 4 | 1,8,10,11 | Spring-Summer | |
| Klebsiella pneumoniae Uber [21] | 1 | 11 | Summer | |
| Salmonella spp. [23] | 1 | 5 | Autumn | |
| Salmonella enterica subsp. enterica serovar Paratyphi A (Ex Kauffmann & Edwards) Le Minor & Popoff [21,23] | 2 | 1,12 | Spring-Winter | |
| Shigella sp. Castellani & Chalmers [21] | 1 | 13 | Spring | |
| Tatumella sp. Hollis et al. [24] | 1 | 13 | Spring | |
| Yersinia enterocolitica (Schleifstein & Coleman) [25] | 1 | 10 | Spring | |
| Pasterurellaceae | Pasteurella multocida Pasteur [26] 2 | 2 | 4,7 | Spring |
| Vibrionaceae | Vibrio cholerae Pacini [21] | 2 | 11,12 | Summer-Autumn |
| Vibro parahemolyticus Fujino et al./Sakazaki et al. [21] | 1 | 8.11 | Summer | |
| Moraxellaceae | Acinetobacter lwoffii Brisou & Prévot [27] | 1 | 6 | Winter |
| Antimicrobial Category | Antimicrobial Agent | Antimicrobial Susceptibility | Species with Intrinsic Resistance to Antimicrobial Agents or Categories (51)a |
|---|---|---|---|
| Aminoglycosides | Gentamicin | S | All isolates |
| Tobramycin | IR | Escherichia coli (1/7) (IR) | |
| Amikacin | R | Escherichia coli (1/7) Klebsiella oxytoca (1/4) (IR) | |
| Antipseudomonal penicillin + β-lactamase inhibitors | Ticarcillin-clavulanic acid | R | Escherichia coli (1/7) |
| Piperacillin-tazobactam (Pip/tazo) | R | Enterobacter cloacae (1/3) Escherichia coli (1/7) Klebsiella oxytoca (1/4) | |
| Carbapenems | Imipenem | IR | Klebsiella oxytoca (1/4) (IR) Tatumella sp. (1/1) (IR) |
| Meropenem | S | All isolates | |
| Non-extended spectrum Cephalosporin: 1st and 2nd generation cephalosporins | Cefazolin | R | Escherichia coli (1/7) Escherichia coli (1/7) (IR) Klebsiella oxytoca (2/4) Klebsiella oxytoca (1/4) (IR) Vibrio parahaemolyicus (1/1) (Vibrionaceae) |
| Cefuroxime | R | Enterobacter cloacae (1/2) Escherichia coli (1/7) Klebsiella oxytoca (1/4) Klebsiella oxytoca (1/4) (IR) Vibrio parahaemolyicus (1/1) (Vibrionaceae) (IR) | |
| Extended-spectrum Cephalosporin: 3rd and 4th generation cephalosporins | Cefotaxime or ceftriaxone | R | Aeromonas hydrophila (1/1) (IR) Citrobacter freundii (1/2) (IR) Enterobacter cloacae (1/3) (IR) Escherichia coli (1/7) Klebsiella oxytoca (3/4) Salmonella enterica subsp. enterica, serovar Paratyphi A (1/2) |
| Ceftazidime | R | Citrobacter freundii (1/2) Enterobacter cloacae (1/3) Escherichia coli (1/7) | |
| Cefepime | R | Escherichia coli (1/7) Enterobacter cloacae (1/3) Klebsiella oxytoca (1/4) Klebsiella oxytoca (1/4) (IR) | |
| Cephamycins | Cefotetan | R | Klebsiella oxytoca (1/4) |
| Fluoroquinolones | Ciprofloxacin | R | Escherichia coli (2/7) Escherichia coli (1/7) (IR) Klebsiella oxytoca (1/4) (IR) |
| Moxifloxacin | R | Escherichia coli (2/7) Escherichia coli (1/7) (IR) Klebsiella oxytoca (1/4) (IR) | |
| Levofloxacin | R | Escherichia coli (2/7) Escherichia coli (1/7) (IR) Klebsiella oxytoca (1/4) S. enterica subsp. enterica, serovar Paratyphi A (1/2) (IR) | |
| Folate pathway inhibitors | Trimethoprim-sulfamethoxazole | R | Aeromonas hydrophila (1/1) Escherichia coli (3/7) |
| Monobactams | Aztreonam | R | Citrobacter freundii (1/2) (IR) Enterobacter cloacae (1/2) Escherichia coli (2/7) Klebsiella oxytoca (1/4) |
| Penicillin | Ampicillin | R | Escherichia coli (2/7) S. enterica subsp. enterica, serovar Paratyphi A (1/2) (IR) Shigella sp. (1/1) Tatumella sp. (1/1) Vibrio cholerae (2/2) (Vibrionaceae) |
| Penicillin + β-lactamase inhibitors | Ampicillin-sulbactam | R | Aeromonas hydrophila (1/1) Enterobacter cloacae (1/3) Escherichia coli (2/7) Hafnia alvei (2/3) (IR) Klebsiella oxytoca (1/4) Shigella sp. (1/1) Tatumella sp. (1/1) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delgado-Gardea, M.C.E.; Tamez-Guerra, P.; Gomez-Flores, R.; Zavala-Díaz de la Serna, F.J.; Eroza-de la Vega, G.; Nevárez-Moorillón, G.V.; Pérez-Recoder, M.C.; Sánchez-Ramírez, B.; González-Horta, M.D.C.; Infante-Ramírez, R. Multidrug-Resistant Bacteria Isolated from Surface Water in Bassaseachic Falls National Park, Mexico. Int. J. Environ. Res. Public Health 2016, 13, 597. https://doi.org/10.3390/ijerph13060597
Delgado-Gardea MCE, Tamez-Guerra P, Gomez-Flores R, Zavala-Díaz de la Serna FJ, Eroza-de la Vega G, Nevárez-Moorillón GV, Pérez-Recoder MC, Sánchez-Ramírez B, González-Horta MDC, Infante-Ramírez R. Multidrug-Resistant Bacteria Isolated from Surface Water in Bassaseachic Falls National Park, Mexico. International Journal of Environmental Research and Public Health. 2016; 13(6):597. https://doi.org/10.3390/ijerph13060597
Chicago/Turabian StyleDelgado-Gardea, Ma. Carmen E., Patricia Tamez-Guerra, Ricardo Gomez-Flores, Francisco Javier Zavala-Díaz de la Serna, Gilberto Eroza-de la Vega, Guadalupe Virginia Nevárez-Moorillón, María Concepción Pérez-Recoder, Blanca Sánchez-Ramírez, María Del Carmen González-Horta, and Rocío Infante-Ramírez. 2016. "Multidrug-Resistant Bacteria Isolated from Surface Water in Bassaseachic Falls National Park, Mexico" International Journal of Environmental Research and Public Health 13, no. 6: 597. https://doi.org/10.3390/ijerph13060597







