Class 1 Integrons and the Antiseptic Resistance Gene (qacEΔ1) in Municipal and Swine Slaughterhouse Wastewater Treatment Plants and Wastewater—Associated Methicillin-Resistant Staphylococcus aureus
Abstract
:1. Introduction
2. Materials and Methods
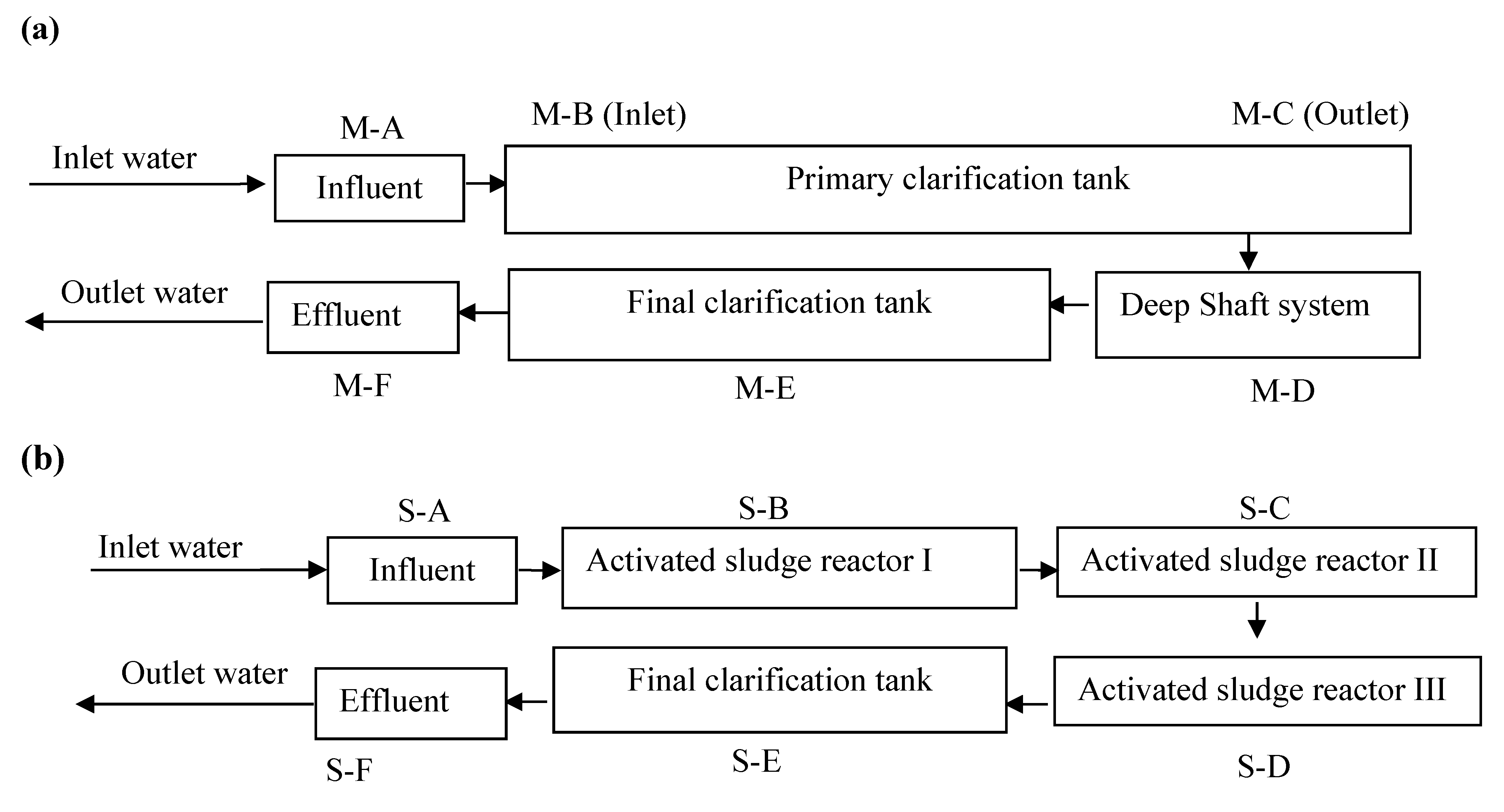
2.1. Wastewater Treatment and Sampling
2.2. Concentration and Total DNA Extraction
2.3. Real-Time qPCR Standards
2.4. Real-Time qPCR
2.5. MRSA Isolation, Identification, Class 1 Integrons and QAC Genes Detection
| Target Genes | Primers | Sequence (5’–3’) | Amplicons (bp) | Annealing Temperature (°C) |
|---|---|---|---|---|
| intI1 | HS463a HS464 | CTGGATTTCGATCACGGCACG ACATGCGTGTAAATCATCGTCG | 473 | 60 |
| qacEΔ1 | qacEΔ1F qacEΔ1B | ATCGCAATAGTTGGCGAAGT CAAGCTTTTGCCCATGAAGC | 225 | 60 |
| nuc | nuc-1 nuc-2 | GCGATTGATGGTGATACGGTI AGCCAAGCCTTGACGAACTAAAGC | 280 | 55 |
| mecA | Forward Reverse | CTCAGGTACTGCTATCCACC CACTTGGTATATCTTCACC | 448 | 42 |
2.6. Minimum Inhibitory Concentration of Antiseptics
2.7. Statistical Analysis
3. Results
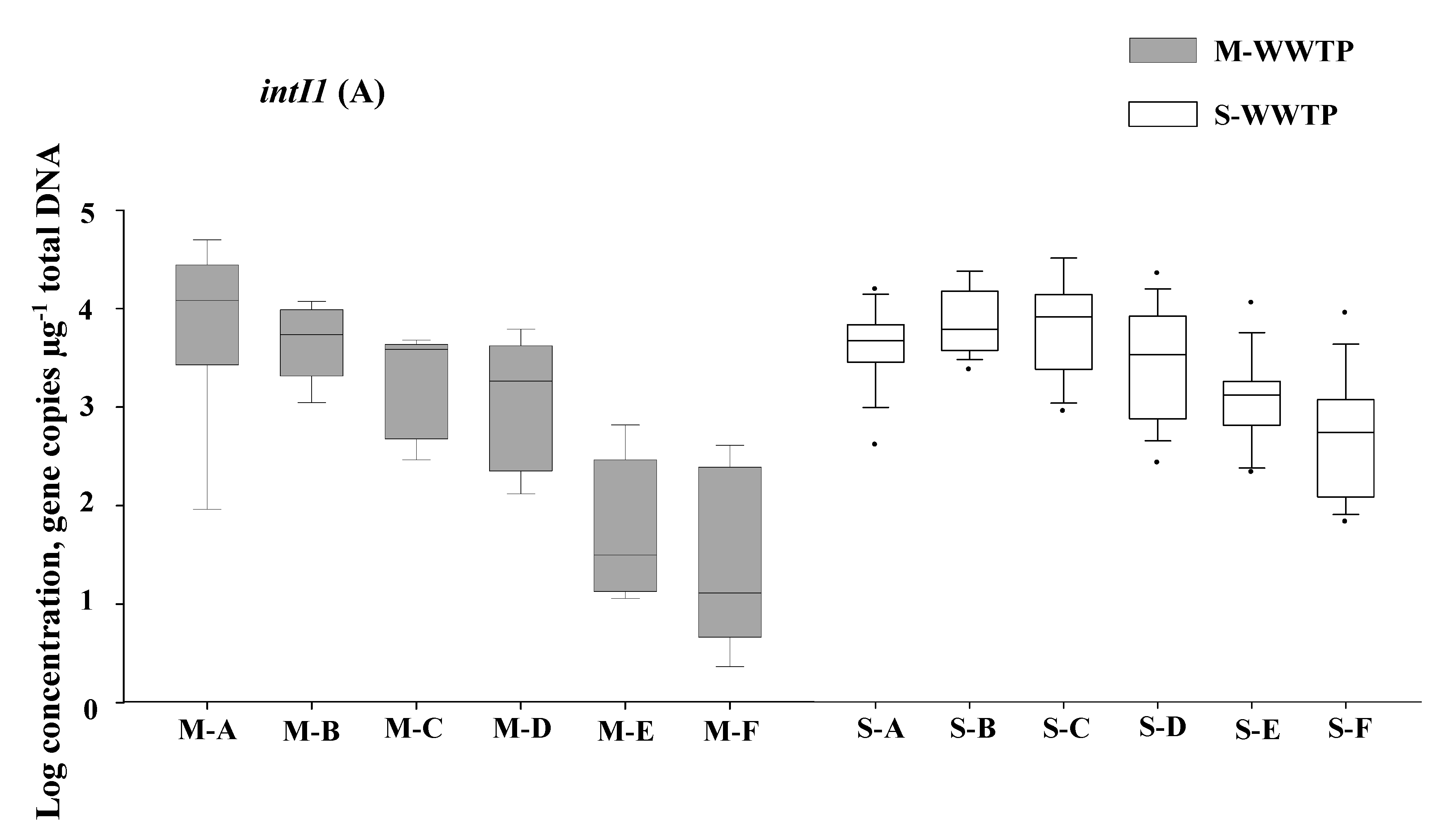
3.1. The Abundances of Inti1 and QacEΔ1 in Wastewater

3.2. Detection of IntI1 and QacEΔ1 in MRSA Isolates and MICs Testing for QacEΔ1-Positive MRSA
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mackie, R.I.; Koike, S.; Krapac, I.; Chee-Sanford, J.; Maxwell, S.; Aminov, R.I. Tetracycline residues and tetracycline resistance genes in groundwater impacted by swine production facilities. Anim. Biotechnol. 2006, 2, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mukherjee, S.; Chakraborty, R. Characterization of a novel trimethoprim resistance gene, dfrA28, in class 1 integron of an oligotrophic Acinetobacter johnsonii strain, MB52, isolated from river Mahananda, India. Microb. Drug Resist. 2010, 16, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Börjesson, S.; Melin, S.; Matussek, A.; Lindgren, P.E. A seasonal study of the mecA gene and Staphylococcus aureus including methicillin-resistant S. aureus in a municipal wastewater treatment plant. Water Res. 2009, 43, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Bouche, H.W.; Corey, G.R. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 2008, 46, S344–S349. [Google Scholar] [CrossRef] [PubMed]
- Harper, A.L.; Ferguson, D.D.; Leedom Larson, K.R.; Hanson, B.M.; Male, M.J.; Donham, K.J.; Smith, T.C. An overview of livestock-associated MRSA in agriculture. J. Agromed. 2010, 15, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, T.; Kohnen, W.; Jansen, B.; Obst, U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 2003, 43, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, R.E.R.; Micallef, S.A.; Gibbs, S.G.; Davis, J.A.; He, X.; George, A.; Kleinfelter, L.M.; Schreiber, N.A.; Mukherjee, S.; Sapkota, A.; et al. Methicillin-resistant Staphylococcus aureus (MRSA) detected at four U.S. wastewater treatment plants. Environ. Health Perspect. 2012, 120, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Stokes, H.W.; Hall, R.M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: Integrons. Mol. Microbiol. 1989, 3, 1669–1683. [Google Scholar] [CrossRef] [PubMed]
- Recchia, G.D.; Hall, R.M. Gene cassettes: A new class of mobile element. Microbiology 1995, 141, 3015–3027. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Z.; Zhang, M.; O’Donoghue, M.; Boost, M. Presence of antiseptic resistance genes in porcine methicillin-resistant Staphylococcus aureus. Vet. Microbiol. 2013, 162, 977–979. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, L.; Shi, L.; Shirtliff, M.E. Class 1 integron in Staphylococci. Mol. Biol. Rep. 2011, 38, 5261–5279. [Google Scholar] [CrossRef] [PubMed]
- Povilonis, J.; Šeputienė, V.; Ružauskas, M.; Šiugždinienė, R.; Virgailis, M.; Pavilonis, A.; Sužiedėlienė, E. Transferable class 1 and 2 Integrons in Escherichia coli and Salmonella enterica isolates of human and animal origin in Lithuania. Foodborne Pathog. Dis. 2010, 7, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, S.A.; Stokes, H.W.; Findlay, S.; Taylor, M.; Gillings, M.R. Quantification of class 1 integron abundance in natural environments using real-time quantitative PCR. FEMS Microbiol. Lett. 2008, 278, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, M. Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from Hangzhou in China. Environ. Sci. Technol. 2013, 47, 8157–8163. [Google Scholar] [PubMed]
- Stokes, H.W.; Nesbo, C.L.; Holley, M.; Bahl, M.I.; Gillings, M.R.; Boucher, Y. Class 1 integrons potentially predating the association with Tn402-like transposition genes are present in a sediment microbial community. J. Bacteriol. 2006, 188, 5722–5730. [Google Scholar] [CrossRef] [PubMed]
- Kücken, D.; Feucht, H.; Kaulfers, P. Association of qacE and qacEDelta1 with multiple resistance to antibiotics and antiseptics in clinical isolates of Gram-negative bacteria. FEMS Microbiol. Lett. 2000, 183, 95–98. [Google Scholar] [CrossRef]
- Noguchi, N.; Nakaminami, H.; Nishijima, S.; Kurokawa, I.; So, H.; Sasatsu, M. Antimicrobial agent of susceptibilities and antiseptic resistance gene distribution among methicillin-resistant Staphylococcus aureus isolates from patients with impetigo and staphylococcal scalded skin syndrome. J. Clin. Microbiol. 2006, 44, 2119–2125. [Google Scholar] [CrossRef] [PubMed]
- Heir, E.; Sundheim, G.; Holck, A.L. The Staphylococcus qacH gene product: A new member of the SMR family encoding multidrug resistance. FEMS Microbiol. Lett. 1998, 163, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Bjorland, J.; Steinum, T.; Kvitle, B.; Waage, S.; Sunde, M.; Heir, E. Widespread distribution of disinfectant resistance genes among staphylococci of bovine and caprine origin in Norway. J. Clin. Microbiol. 2005, 43, 4363–4368. [Google Scholar]
- Raggi, C.; Filippini, P.; Monaco, M.; Pantosti, A.; Creti, R.; Baldassarri, L. Methicillin resistance, biofilm formation and resistance to benzalkonium chloride in Staphylococcus aureus clinical isolates. Clin. Microbial. 2013. [Google Scholar] [CrossRef]
- Wan, M.T.; Chou, C.C. Spreading of β-lactam resistance gene (mecA) and methicillin-resistant Staphylococcus aureus through municipal and swine slaughterhouse wastewaters. Water Res. 2014, 64, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Sandvang, D.; Aarestrup, F.M.; Jensen, L.B. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 1997, 157, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS ONE 2011. [Google Scholar] [CrossRef]
- Brakstad, O.G.; Aasbakk, K.; Maeland, J.A. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 1992, 30, 1654–1660. [Google Scholar] [PubMed]
- Sakoulas, G.; Gold, H.S.; Venkataraman, L.; DeGirolami, P.C.; Eliopoulos, G.M.; Qian, Q. Methicillin-resistant Staphylococcus aureus: Comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J. Clin. Microbiol. 2001, 39, 3946–3951. [Google Scholar] [CrossRef] [PubMed]
- Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2003.
- Jacobs, L.; Chenia, H.Y. Characterization of integrons and tetracycline resistance determinants in Aeromonas spp. isolated from South African aquaculture systems. Int. J. Food Microbiol. 2007, 114, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Taviani, E.; Ceccarelli, D.; Lazaro, N.; Bani, S.; Cappuccinelli, P.; Colwell, R.R.; Colombo, M.M. Environmental Vibrio spp., isolated in Mozambique, contain a polymorphic group of integrative conjugative elements and class 1 integrons. FEMS Microbiol. Ecol. 2008, 64, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.C.; Chiu, T.H.; Pang, J.C.; Hsuan-Yuan, C.H.; Chang, G.N.; Tsen, H.Y. Characterisation of antimicrobial resistance patterns and class 1 integrons among Escherichia coli and Salmonella enterica serovar Choleraesuis strains isolated from humans and swine in Taiwan. Int. J. Antimicrob. Agents 2006, 27, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Betteridge, T.; Partridge, S.R.; Iredell, J.R.; Stokes, H.W. Genetic context and structural diversity of class 1 integrons from human commensal bacteria in a hospital intensive care unit. Antimicrob. Agents Chemother. 2011, 55, 3939–3943. [Google Scholar] [CrossRef] [PubMed]
- Gaze, W.H.; Abdouslam, N.; Hawkey, P.M.; Wellington, E.M. Incidence of class 1 integrons in a quaternary ammonium compound-polluted environment. Antimicrob. Agents Chemother. 2005, 49, 1802–1807. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.; Boucher, Y.; Labbate, M.; Holmes, A.; Krishnan, S.S.; Holley, M.; Stokes, H.W. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 2008, 190, 5095–5100. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.M.; Li, C.Y.; Ho, M.W.; Lin, C.Y.; Liu, S.H.; Lu, J.J. High rate of qacA- and qacB-positive methicillin-resistant Staphylococcus aureus isolates from chlorhexidine-impregnated catheter-related bloodstream infections. Antimicrob. Agents Chemother. 2012, 56, 5693–5697. [Google Scholar] [CrossRef] [PubMed]
- Mayer, S.; Boos, M.; Beyer, A.; Fluit, A.C.; Schmitz, F.J. Distribution of the antiseptic resistance genes qacA, qacB and qacC in 497 methicillin-resistant and—Susceptible European isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 2001, 47, 896–897. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, M.T.; Chou, C.C. Class 1 Integrons and the Antiseptic Resistance Gene (qacEΔ1) in Municipal and Swine Slaughterhouse Wastewater Treatment Plants and Wastewater—Associated Methicillin-Resistant Staphylococcus aureus. Int. J. Environ. Res. Public Health 2015, 12, 6249-6260. https://doi.org/10.3390/ijerph120606249
Wan MT, Chou CC. Class 1 Integrons and the Antiseptic Resistance Gene (qacEΔ1) in Municipal and Swine Slaughterhouse Wastewater Treatment Plants and Wastewater—Associated Methicillin-Resistant Staphylococcus aureus. International Journal of Environmental Research and Public Health. 2015; 12(6):6249-6260. https://doi.org/10.3390/ijerph120606249
Chicago/Turabian StyleWan, Min Tao, and Chin Cheng Chou. 2015. "Class 1 Integrons and the Antiseptic Resistance Gene (qacEΔ1) in Municipal and Swine Slaughterhouse Wastewater Treatment Plants and Wastewater—Associated Methicillin-Resistant Staphylococcus aureus" International Journal of Environmental Research and Public Health 12, no. 6: 6249-6260. https://doi.org/10.3390/ijerph120606249





