Resistance of Stenotrophomonas maltophilia to Fluoroquinolones: Prevalence in a University Hospital and Possible Mechanisms
Abstract
:1.Introduction
2. Materials and Methods
2.1. S. maltophilia Isolates
2.2. Determination of Minimum Inhibitory Concentration of Fluoroquinolones
2.3. Effect of Reserpine on Fluoroquinolone MIC
2.4. Detection of Fluoroquinolone Resistance Genes in S. maltophilia Clinical Isolates
| Fluoroquinolone Resistance Gene | Sequence | Annealing Temperature (°C) | Product Size (bp) |
|---|---|---|---|
| qnrA | F: 5’-AGAGGATTTCTCACGCCAGG-3’; R: 5’-TGCCAGGCACAGATCTTGAC-3’ | 63 | 580 |
| qnrB | F: 5’-GGMATHGAAATTCGCCACTG-3’; R: 5’- TTTGCYGYYCGCCAGTCGAA | 56 | 264 |
| qnrS | F: 5’-GCAAGTTCATTGAACAGGGT-3’; R: 5’-TCTAAACCGTCGAGTTCGGCG-3’ | 56 | 428 |
| qnrC | F: 5’-GGGTTGTACATTTATTGAATCG-3’; R: 5’-CACCTACCCATTTATTTTCA-3’ | 56 | 310 |
| qnrD | F: 5’-GGGTTGATTTAACTGATAC-3’; R: 5’-TTCGCACTTTTCTAATATGAC-3’ | 56 | 310 |
| aac-Ib-Cr | F: 5’-TTGCGATGCTCTATGAGTGGCTA-3’; R: 5’-CTCGAATGCCTGGCGTGTTT-3’ | 58 | 482 |
| qepA | F: 5’-GCAGGTCCAGCAGCGGGTAG-3’; R: 5’-CTTCCTGCCCGAGTATCGTG-3’ | 48 | 199 |
| oqxA | F: 5’-CTTGCACTTAGTTAAGCGCC-3’; R: 5’-GAGGTTTTGATAGTGGAGGTAGG-3’ | 65 | 866 |
| oqxB | F: 5’-GCGGTGCTGTCGATTTTA-3’; R: 5’- TACCGGAACCCATCTCGAT-3’ | 65 | 781 |
| smeE | F: 5’-AGCTCGACGCCACGGTA-3’; R: 5’- TGGCCTGGATCGAGAGCA-3’ | 55 | 803 |
| smeF | F: 5’-GCCACGCTGAAGACCTA-3’; R: 5’- CACCTTGTACAGGGTGA-3’ | 55 | 800 |
| smeD | F: 5’-CCAAGAGCCTTTCCGTCAT-3’; R: 5’- TCTCGGACTTCAGCGTGAC-3’ | 58 | 150 |
| gyrA | F: 5’-AACTCAACGCGCACAGCAACAAGCC-3’; R: 5’-CCAGTTCCTTTTCGTCGTAGTTGGG-3’ | 58 | 300 |
| parC | F: 5’-ATCGGCGACGGCCTGAAGCC-3’; R: 5’-CGGGATTCGGTATAACGCAT-3’ | 55 | 273 |
| smqnr | F: 5’-GCTCTAGAGCTCTACGAATGCGATTTCTCCG-3’; R: 5’- CGGAATTCCGAAACTGGCACCGCTCACG-3’ | 52 | 817 |
| gyrA* | F: 5’-AACTCAACGCGCACAGCAACAAGCC-3’; R: 5’-CCAGTTCCTTTTCGTCGTAGTTGGG-3’ | 58 | 300 |
| smeD* | F: 5’-CCAAGAGCCTTTCCGTCAT-3’; R: 5’- TCTCGGACTTCAGCGTGAC-3’ | 58 | 150 |
| smeF* | F: 5’-CCAACGCGGATCGTGATATC-3’; R: 5’-TGCTCATCCAGGCTGACATTC-3’ | 58 | 100 |
2.5. Quantitative Fluorescent (QF)-PCR
2.6. PFGE
2.7. Statistics
3. Results
3.1. Prevalence of S. maltophilia clinical Isolates
3.2. Resistance of S. maltophilia to Fluoroquinolones
3.3. Fluoroquinolone Resistance in S. maltophilia Isolated from Various Departments of the Hospital
| Fluoroquinolone Antibacterial | 2010 (n = 69) | 2011 (n = 148) | 2012 (n = 209) | χ2 value | p value | |||
|---|---|---|---|---|---|---|---|---|
| Resistant (%) | Susceptible (%) | Resistant (%) | Susceptible (%) | Resistant (%) | Susceptible (%) | |||
| Levofloxacin | 0 | 100 | 4 | 94.6 | 4.1 | 93 | 3.102 | 0.212 |
| Sulfamethoxazole | 53 | 47 | 66 | 34 | 86.6 | 13.4 | 36.963 | 0 |
| Minocycline | 0 | 100 | 0.5 | 99 | 0.7 | 98 | 0.695 | 0.706 |
| Department | No. Bacterial Isolate | Levofloxacin | Sulfamethoxazole | Minocycline | |||
|---|---|---|---|---|---|---|---|
| Resistant (%) | Susceptible (%) | Resistant (%) | Susceptible (%) | Resistant (%) | Susceptible (%) | ||
| ICU | 113 | 2.7 | 97.3 | 72.1 | 27.9 | 0.9 | 98.2 |
| Respiratory medicine | 82 | 6.3 | 92.4 | 72.2 | 27.8 | 1.3 | 98.7 |
| Neurosurgery | 49 | 8.5 | 89.4 | 82.6 | 17.4 | 0 | 97.9 |
| Pediatrics | 43 | 0 | 97.7 | 72.1 | 27.9 | 0 | 100 |
| Emergency | 29 | 0 | 89.3 | 75 | 25 | 0 | 100 |
| Neurology | 20 | 0 | 95 | 95 | 5 | 0 | 100 |
| Others | 90 | 12.4 | 86.2 | 81.7 | 18.3 | 0 | 98.6 |
| Age Group (years) | Bacterial Isolate | Levofloxacin | Sulfamethoxazole | Minocycline | ||||
|---|---|---|---|---|---|---|---|---|
| No. | Percentage (%) | Resistant (%) | Susceptible (%) | Resistant (%) | Susceptible (%) | Resistant (%) | Susceptible (%) | |
| <18 | 59 | 13.8 | 0 | 98.6 | 76.8 | 23.2 | 0 | 100 |
| 18–59 | 129 | 30.3 | 2.1 | 96.8 | 70.6 | 29.4 | 0.5 | 98.9 |
| ≥60 | 238 | 55.9 | 6.3 | 91.2 | 77.1 | 22.9 | 0.6 | 98.2 |
| Antimicrobial | MICR (µg/mL) | MIC50 (µg/mL) | MIC90 (µg/mL) | Percentage of Susceptible Isolate (%) | Percentage of Intermediate Isolate (%) | Percentage of Resistant Isolate (%) |
|---|---|---|---|---|---|---|
| Norfloxacin | 0.5–128 | 8 | 64 | 45.7 | 21.9 | 32.4 |
| Ofloxacin | 0.125–64 | 1 | 4 | 73.6 | 13.2 | 13.2 |
| Chloramphenicol | 0.25–64 | 8 | 64 | 51.7 | 23.7 | 24.6 |
| Minocycline | 0.25–64 | 1 | 4 | 90.3 | 4.4 | 5.3 |
| Ceftazidime | 0.5–128 | 8 | 128 | 66.7 | 10.5 | 22.8 |
| Levofloxacin | 0.25–64 | 2 | 8 | 73.7 | 14.0 | 12.3 |
| Ciprofloxacin | 0.25–64 | 1 | 4 | 54.4 | 23.7 | 21.9 |
| Antimicrobial | MICR (µg/mL) | No. Resistant Isolate | Percentage of Resistant Isolate (%) | No. Intermediate Isolate | Percentage of Intermediate Isolate (%) | No. Susceptible Isolate | Percentage of Susceptible Isolate (%) |
|---|---|---|---|---|---|---|---|
| Norfloxacin | 0.5–128 | 37 | 32.4 | 25 | 21.9 | 52 | 45.7 |
| Norfloxacin +reserpine | 0.5–128 | 25 | 21.9 | 19 | 16.7 | 70 | 61.4 |
| Ciprofloxacin | 0.25–64 | 25 | 21.9 | 27 | 23.7 | 62 | 54.4 |
| Ciprofloxacin + reserpine | 0.25–64 | 15 | 13.2 | 22 | 19.3 | 77 | 67.5 |
| Ofloxacin | 0.125–64 | 15 | 13.2 | 15 | 13.2 | 84 | 73.6 |
| Ofloxacin + reserpine | 0.125–64 | 11 | 9.6 | 11 | 9.6 | 92 | 80.8 |
| Antimicrobial | Efflux Pump-Positive Isolate (n = 19) | Efflux Pump-Negative Isolate (n = 95) | ||||
|---|---|---|---|---|---|---|
| Resistant (%) | Intermediate (%) | Susceptible (%) | Resistant (%) | Intermediate (%) | Susceptible (%) | |
| Norfloxacin | 18 (94.7) | 0 | 1 (5.3) | 19 (20) | 25 (26.3) | 51 (53.7) |
| Ciprofloxacin | 16 (94.2) | 1 (5.3) | 2 (10.5) | 9 (9.5) | 26 (27.4) | 60 (63.1) |
| Ofloxacin | 15 (78.9) | 3 (15.8) | 1 (5.3) | 0 | 12 (12.6) | 83 (87.4) |
3.4. Fluoroquinolone Resistance in S. maltophilia Isolated from Patients at Various Age Groups
3.5. Effect of Reserpine on MIC of S. maltophilia to Fluoroquinolone
3.6. Association of Efflux Pump Phenotype with Resistance of S. maltophilia to Fluoroquinolone
3.7. Prevalence of Fluoroquinolone Resistance Genes
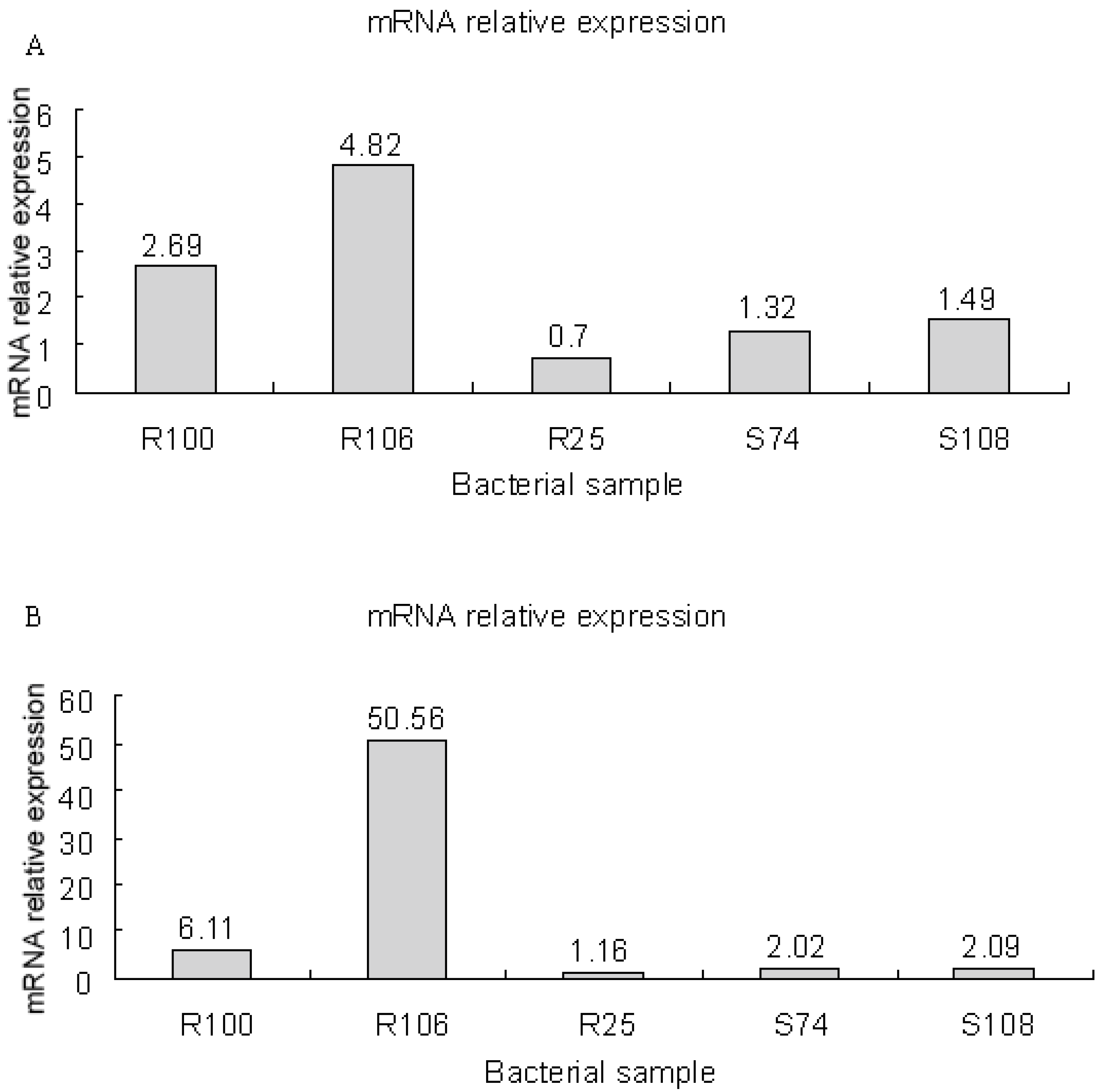
3.8. mRNA Expression of smeD and smeF in S. maltophilia
3.9. Sequence Analysis of gyrA, parC and smqnr Genes
3.10. PFGE
4. Discussion
| Bacteria Number | Antimicrobial MIC (µg/mL) | gyrA | parC | |||||
|---|---|---|---|---|---|---|---|---|
| Norfloxacin | Ciprofloxacin | Ofloxacin | 85th amino acid | 119th amino acid | 151th amino acid | 37th amino acid | 72th amino acid | |
| ATCC13637 | 0.5 | 0.25 | 0.25 | Valine (GTC) | Alanine (GCA) | Glutamic acid (GAA) | Glycin (GGC) | Glycin (GGT) |
| S31 | 4 | 0.5 | 0.5 | Valine (GTG) | Alanine (GCG) | — | Arginine (CGC) | — |
| S62 | 2 | 1 | 1 | Valine (GTG) | Alanine (GCG) | Lysine (AAA) | — | — |
| R101 | 32 | 8 | 16 | Valine (GTG) | Alanine (GCG) | Lysine (AAA) | Arginine (CGC) | — |
| S126 | 8 | 2 | 2 | Valine (GTG) | Alanine (GCG) | — | Arginine (CGC) | Glycin (GGC) |
| R138 | 32 | 4 | 8 | Valine (GTG) | — | Lysine (AAA) | Arginine (CGC) | Glycin (GGC) |

5. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Looney, W.J.; Narita, M.; Mühlemann, K. Stenotrophomonas maltophilia: An emerging opportunist human pathogen. Lancet Infect. Dis. 2009, 9, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin.Microbiol. Res. 2012, 25, 2–41. [Google Scholar] [CrossRef]
- Palleroni, N.J.; Bradbury, J.F. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int. J. Syst. Bacteriol. 1993, 43, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Cheong, H.S.; Lee, J.A.; Kang, C.I.; Chung, D.R.; Peck, K.R.; Kim, E.S.; Lee, J.S.; Son, J.S.; Lee, N.Y.; Song, J.H. Risk factors for mortality and clinical implications of catheter-related infections in patients with bacteraemia caused by Stenotrophomonas maltophilia. Int. J.Antimicrob. Agents 2008, 32, 538–540. [Google Scholar] [CrossRef] [PubMed]
- Kara, I.H.; Yilmaz, M.E.; Sit, D.; Kadiroğ, A.K.; Kökoğlu, O.F. Bacteremia caused by Stenotrophomonas maltophilia in a dialysis patient with a long-term central venous catheter. Infect. Control Hosp. Epidemiol. 2006, 27, 535–536. [Google Scholar] [CrossRef] [PubMed]
- Abbott, I.J.; Slavin, M.A.; Turnidge, J.D.; Thursky, K.A.; Worth, L.J. Stenotrophomonas maltophilia: Emerging disease patterns and challenges for treatment. Expert Rev. Anti Infect.Ther. 2011, 9, 471–488. [Google Scholar] [CrossRef] [PubMed]
- Nicodemo, A.C.; Paez, J.I. Antimicrobial therapy for Stenotrophomonas maltophilia infections. Eur. J.Clin. Microbiol. Infect. Dis. 2007, 26, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Waters, V. New treatments for emerging cystic fibrosis pathogens other than Pseudomonas. Curr. Pharm. Des. 2012, 18, 696–725. [Google Scholar] [CrossRef] [PubMed]
- Bellido, J.L.; Hernández, F.J.; Zufiaurre, M.N.; García-Rodrí, J.A. In vitro activity of newer fluoroquinolones against Stenotrophomonas maltophilia. J.Antimicrob.Chemother. 2000, 46, 334–335. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, F.J.; Verhoef, J.; Fluit, A.C. Comparative activities of six different fluoroquinolones against 9,682 clinical bacterial isolates from 20 European university hospitals participating in the European SENTRY surveillance programme. The SENTRY participants group. Int. J. Antimicrob. Agents 1999, 12, 311–317. [Google Scholar] [CrossRef]
- Isenberg, H.D.; Alperstein, P.; France, K. In vitro activity of ciprofloxacin, levofloxacin, and trovafloxacin, alone and in combination with beta-lactams, against clinical isolates of Pseudomonas aeruginosa, Stenotrophomonas maltophilia, and Burkholderia cepacia. Diagn.Microbiol. Infect. Dis. 1999, 33, 81–86. [Google Scholar] [CrossRef]
- TrigoDaporta, M.; Muñoz Bellido, J.L.; García-Rodríguez, J.A. Topoisomerases mutations and fluoroquinolone resistance in Stenotrophomonas maltophilia. Int. J. Antimicrob. Agents 2004, 24, 520–521. [Google Scholar] [CrossRef]
- Garrison, M.W.; Anderson, D.E.; Campbell, D.M.; Carroll, K.C.; Malone, C.L.; Anderson, J.D.; Hollis, R.J.; Pfaller, M.A. Stenotrophomonas maltophilia: Emergence of multidrug-resistant strains during therapy and in an in vitro pharmacodynamic chamber model. Antimicrob. Agents Chemother. 1996, 40, 2859–2864. [Google Scholar]
- Baek, J.H.; Kim, C.O.; Jeong, S.J.; Ku, N.S.; Han, S.H.; Choi, J.Y.; Yong, D.; Song, Y.G.; Lee, K.; Kim, J.M. Clinical factors associated with acquisition of resistance to levofloxacin in Stenotrophomonas maltophilia. Yonsei Med. J. 2014, 55, 987–993. [Google Scholar] [CrossRef]
- Oizumi, N.; Kawabata, S.; Hirao, M.; Watanabe, K.; Okuno, S.; Fujiwara, T.; Kikuchi, M. Relationship between mutations in the DNA gyrase and topoisomerase IV genes and nadifloxacin resistance in clinically isolated quinolone-resistant Staphylococcus aureus. J. Infect.Chemother. 2001, 7, 191–194. [Google Scholar] [CrossRef]
- Drlica, K.; Zhao, X. DNA gyrase, topoisomerase IV,and the 4-quinolones. Microbiol. Mol. Biol. Rev. 1997, 61, 377–392. [Google Scholar]
- Vranakis, I.; Sandalakis, V.; Chochlakis, D.; Tselentis, Y.; Psaroulaki, A. DNA gyrase and topoisomerase IV mutations in an in vitro fluoroquinolone-resistant Coxiella burnetii strain. Microb. Drug Resist. 2010, 16, 111–117. [Google Scholar] [CrossRef]
- Weigel, L.M.; Anderson, G.J.; Tenover, F.C. DNA gyrase and topoisomerase IV mutations associated with fluoroquinolone resistance in Proteus mirabilis. Antimicrob. Agents Chemother. 2002, 46, 2582–2587. [Google Scholar] [CrossRef]
- García-León, G.; Salgado, F.; Oliveros, J.C.; Sánchez, M.B.; Martínez, J.L. Interplay between intrinsic and acquired resistance to quinolones in Stenotrophomonas maltophilia. Environ.Microbiol. 2014, 16, 1282–1296. [Google Scholar] [CrossRef]
- Gordon, N.C.; Wareham, D.W. Novel variants of the Smqnr family of quinolone resistance genes in clinical isolates of Stenotrophomonas maltophilia. J.Antimicrob.Chemother. 2010, 65, 483–489. [Google Scholar] [CrossRef]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef]
- Poole, K. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 2001, 3, 255–264. [Google Scholar]
- Tsaur, S.M.; Chang, S.C.; Luh, K.T.; Hsieh, W.C. Antimicrobial susceptibility of enterococci in vitro. J. Formos. Med. Assoc. 1993, 92, 547–552. [Google Scholar]
- Valdezate, S.; Vindel, A.; Echeita, A.; Baquero, F.; Cantó, R. Topoisomerase II and IV quinolone resistance-determining regions in Stenotrophomonas maltophilia clinical isolates with different levels of quinolone susceptibility. Antimicrob. Agents Chemother. 2002, 46, 665–671. [Google Scholar] [CrossRef]
- Sun, E.L.; Song, S.D.; Wei, D.J. Effect of efflux inhibitor on the activity of fluoroquinlone to Stenotrophomonas maltophilia. Chin. J.Antibiot. 2003, 28, 757–760. [Google Scholar]
- Zhao, J.Y.; Dang, H. Coastal seawater bacteria harbor a large reservoir of plasmid-mediated quinolone resistance determinants in Jiaozhou Bay, China. Microb. Ecol. 2012, 64, 187–199. [Google Scholar] [CrossRef]
- Gould, V.C.; Avison, M.B. SmeDEF-mediated antimicrobial drug resistance in Stenotrophomonas maltophilia clinical isolates having defined phylogenetic relationships. J. Antimicrob. Chemother. 2006, 57, 1070–1076. [Google Scholar] [CrossRef]
- Yuan, J.; Xu, X.; Guo, Q.; Zhao, X.; Ye, X.; Guo, Y.; Wang, M. Prevalence of the oqxAB gene complex in Klebsiella pneumonia and Escherichia coli clinical isolates. J.Antimicrob. Chemother. 2012, 67, 1655–1659. [Google Scholar] [CrossRef]
- Zhang, R.; Sun, Q.; Hu, Y.J.; Yu, H.; Li, Y.; Shen, Q.; Li, G.X.; Cao, J.M.; Yang, W.; Wang, Q.; et al. Detection of the Smqnr quinolone protection gene and its prevalence in clinical isolates of Stenotrophomonas maltophilia in China. J. Med. Microbiol. 2012, 61, 535–539. [Google Scholar] [CrossRef]
- Alonso, A.; Martínez, J.L. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2000, 44, 3079–3086. [Google Scholar] [CrossRef]
- Chang, L.L.; Chen, H.F.; Chang, C.Y.; Lee, T.M.; Wu, W.J. Contribution of integrons, and SmeABC and SmeDEF efflux pumps to multidrug resistance in clinical isolates of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2004, 53, 518–521. [Google Scholar] [CrossRef]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J.Clin.Microbiol. 1995, 33, 2233–2239. [Google Scholar]
- Falagas, M.E.; Valkimadi, P.E.; Huang, Y.T.; Matthaiou, D.K.; Hsueh, P.R. Therapeutic options for Stenotrophomonas maltophilia infections beyond co-trimoxazole: A systematic review. J. Antimicrob. Chemother. 2008, 62, 889–894. [Google Scholar] [CrossRef]
- Ughachukwu, P.O.; Unekwe, P.C. Efflux pump-mediated resistance in chemotherapy. Ann. Med. Health Sci. Res. 2012, 2, 191–198. [Google Scholar] [CrossRef]
- Gibbons, S.; Udo, E.E. The effect of reserpine, a modulator of multidrug efflux pumps, on the in vitro activity of tetracycline against clinical isolates of methicillin resistant Staphylococcus aureus (MRSA) possessing the tet(K) determinant. Phytother. Res. 2000, 14, 139–140. [Google Scholar]
- Schmitz, F.J.; Fluit, A.C.; Lückefahr, M.; Engler, B.; Hofmann, B.; Verhoef, J.; Heinz, H.P.; Hadding, U.; Jones, M.E. The effect of reserpine, an inhibitor of multidrug efflux pumps, on the in vitro activities of ciprofloxacin, sparfloxacin and moxifloxacin against clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 1998, 42, 807–810. [Google Scholar] [CrossRef]
- Li, X.Z.; Zhang, L.; Poole, K. SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2002, 46, 333–343. [Google Scholar]
- Zhang, L.; Li, X.Z.; Poole, K. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2001, 45, 3497–3503. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Nikaido, H. Efflux-mediated drug resistance in bacteria: An update. Drugs 2009, 69, 1555–1623. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Martinez, J.L. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2001, 45, 1879–1881. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.L.; Song, S.D.; Qi, W.; Guo, Q.L. Stenotrophomonas maltophilia SmeDEF efflux pump and the regulation mechanism. Chin. J.Microbiol.Immunol. 2004, 21, 743–747. [Google Scholar]
- Zhao, X.; Xu, C.; Domagala, J.; Drlica, K. DNA topoisomerase targets of the fluoroquinolones: A strategy for avoiding bacterial resistance. Proc. Natl. Acad. Sci. USA 1997, 94, 13991–13996. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.R.; Malik, M.; Snyder, M.; Drlica, K. DNA gyrase and topoisomerase IV on the bacterial chromosome: Quinolone-induced DNA cleavage. J. Mol. Biol. 1996, 258, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Bearden, D.T.; Danziger, L.H. Mechanism of action of and resistance to quinolones. Pharmacotherapy 2001, 21, S224–S232. [Google Scholar] [CrossRef]
- Reece, R.J.; Maxwell, A. DNA gyrase: Structure and function. Crit. Rev.Biochem. Mol. Biol. 1991, 26, 335–375. [Google Scholar] [CrossRef] [PubMed]
- Weigel, L.M.; Steward, C.D.; Tenover, F.C. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 1998, 42, 2661–2667. [Google Scholar] [PubMed]
- Ribera, A.; Doménech-Sanchez, A.; Ruiz, J.; Benedi, V.J.; Jimenez de Anta, M.T.; Vila, J. Mutations in gyrA and parC QRDRs are not relevant for quinolone resistance in epidemiological unrelated Stenotrophomonas maltophilia clinical isolates. Microb. Drug Resist. 2002, 8, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, Z.X.; Yao, J.; Luo, B. Relationship between gyrA and parC gene and Stenotrophomonas maltophilia to fluoroquinolone resistance. Acta Univ. Med. Anhui 2011, 46, 209–212. [Google Scholar]
- Wang, M.; Sahm, D.F.; Jacoby, G.A.; Hooper, D.C. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob. Agents Chemother. 2004, 48, 1295–1299. [Google Scholar] [CrossRef] [PubMed]
- Silva-Sánchez, J.; Cruz-Trujillo, E.; Barrios, H.; Reyna-Flores, F.; Sánchez-Pérez, A.; Bacterial Resistance Consortium; Garza-Ramos, U. Characterization of plasmid-mediated quinolone resistance (PMQR) genes in extended-spectrum β-lactamase-producing Enterobacteriaceae pediatric clinical isolates in Mexico. PLoS ONE 2013, 8, e77968. [Google Scholar] [CrossRef] [PubMed]
- Cruz, G.R.; Radice, M.; Sennati, S.; Pallecchi, L.; Rossolini, G.M.; Gutkind, G.; Conza, J.A. Prevalence of plasmid-mediated quinolone resistance determinants among oxyimino cephalosporin-resistant Enterobacteriaceae in Argentina. Mem. Inst. Oswaldo Cruz 2013, 108, 924–927. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Yu, T.; Jiang, X.; Zhang, W.; Zhang, L.; Ma, J. Emergence of plasmid-mediated quinolone resistance genes in clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa in Henan, China. Diagn. Microbiol. Infect. Dis. 2014, 79, 381–383. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.B.; Martínez, J.L. SmQnr contributes to intrinsic resistance to quinolones in Stenotrophomonas maltophilia. Antimicrob. Agents. Chemother. 2010, 54, 580–581. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Kikuchi, K.; Sasaki, T.; Takahashi, N.; Ohtsuka, M.; Ono, Y.; Hiramatsu, K. Smqnr, a new chromosome-carried quinolone resistance gene in Stenotrophomonas maltophilia. Antimicrob. Agents. Chemother. 2008, 52, 3823–3825. [Google Scholar] [CrossRef] [PubMed]
- Goering, R.V. Pulsed field gel electrophoresis: A review of application and interpretation in the molecular epidemiology of infectious disease. Infect. Genet. Evol. 2010, 10, 866–875. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, W.; Wang, J.; Xu, H.; Li, G. Resistance of Stenotrophomonas maltophilia to Fluoroquinolones: Prevalence in a University Hospital and Possible Mechanisms. Int. J. Environ. Res. Public Health 2015, 12, 5177-5195. https://doi.org/10.3390/ijerph120505177
Jia W, Wang J, Xu H, Li G. Resistance of Stenotrophomonas maltophilia to Fluoroquinolones: Prevalence in a University Hospital and Possible Mechanisms. International Journal of Environmental Research and Public Health. 2015; 12(5):5177-5195. https://doi.org/10.3390/ijerph120505177
Chicago/Turabian StyleJia, Wei, Jiayuan Wang, Haotong Xu, and Gang Li. 2015. "Resistance of Stenotrophomonas maltophilia to Fluoroquinolones: Prevalence in a University Hospital and Possible Mechanisms" International Journal of Environmental Research and Public Health 12, no. 5: 5177-5195. https://doi.org/10.3390/ijerph120505177





