Biodegradation of di-n-Butyl Phthalate by Achromobacter sp. Isolated from Rural Domestic Wastewater
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals
2.2. Isolation and Identification of Bacteria
2.3. Substrate Utilization Tests
2.4. Effects of pH, Temperature, and Agitation Rate on DBP Biodegradation
2.5. Effects of Heavy Metals and Surfactant on DBP Degradation by Strain W-1
2.6. Kinetics of DBP Degradation by Strain W-1
2.7. Plasmid Eliminate Experiment
2.8. Analysis of DBP Residue
3. Results and Discussion
3.1. Isolation and Identification of Bacteria


3.2. Substrate Utilization Tests
| Substrate | DMP/OD600 | DEP/OD600 | DBP/OD600 | DEHP/OD600 | DOP/OD600 | Phenol/OD600 |
|---|---|---|---|---|---|---|
| Utilization | +/0.340 | +/0.426 | +/0.537 | +/1.179 | +/0.962 | +/0.902 |
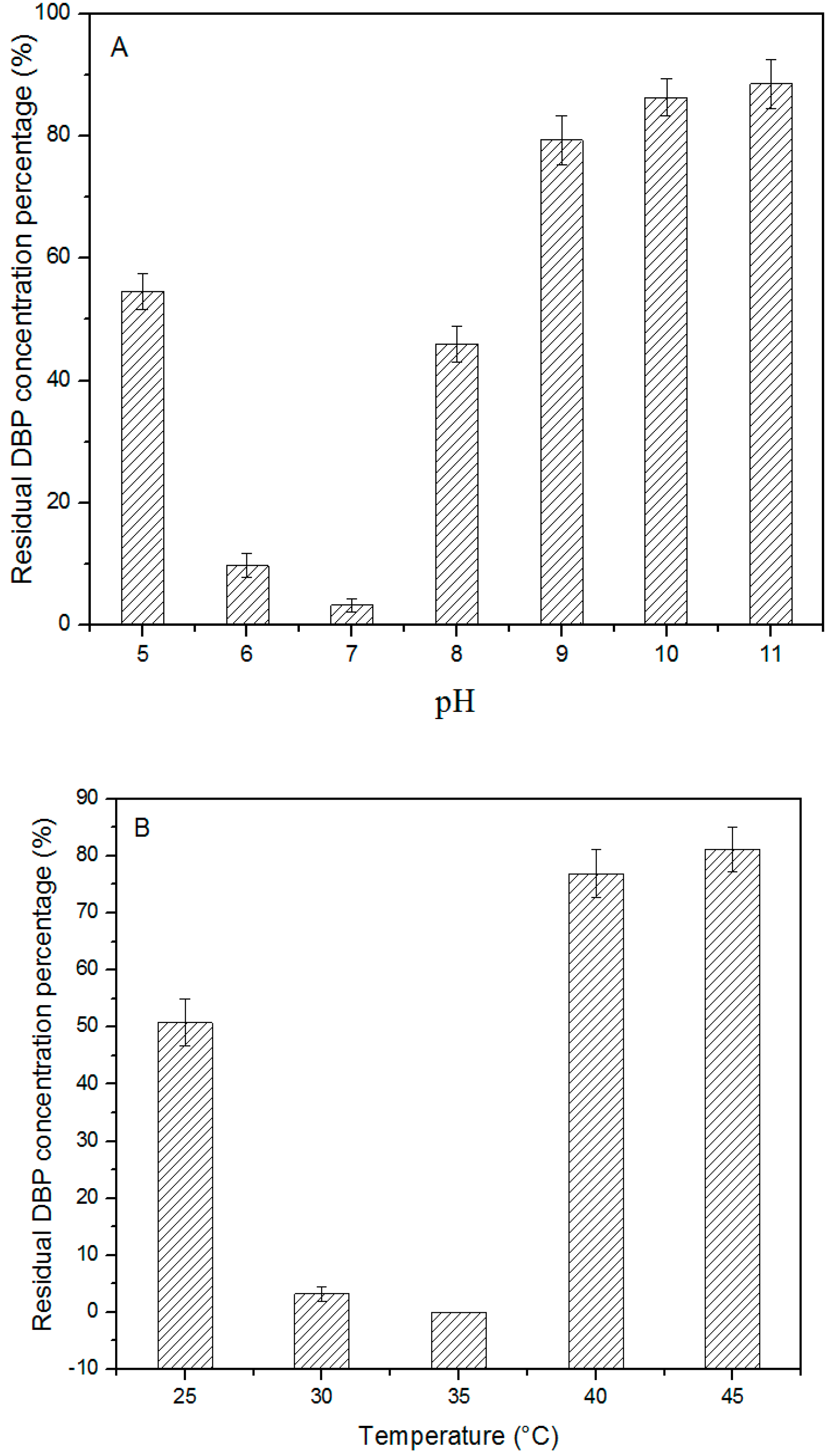
3.3. Effects of pH, Temperature, and Agitation rate on DBP Biodegradation

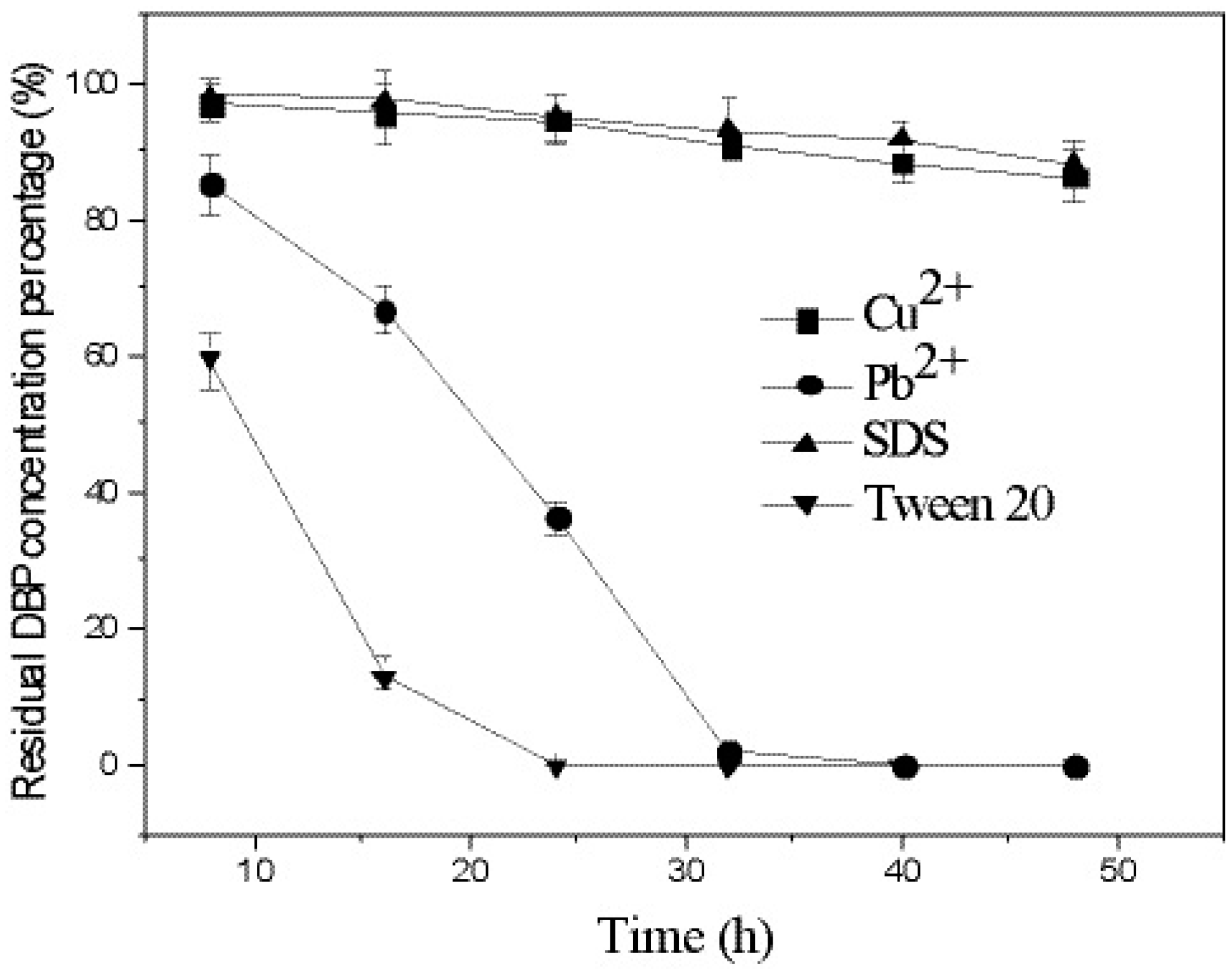
3.4. Effects of Heavy Metals and Surfactants on DBP Degradation by Strain W-1
3.5. Kinetics of DBP Degradation by Strain W-1
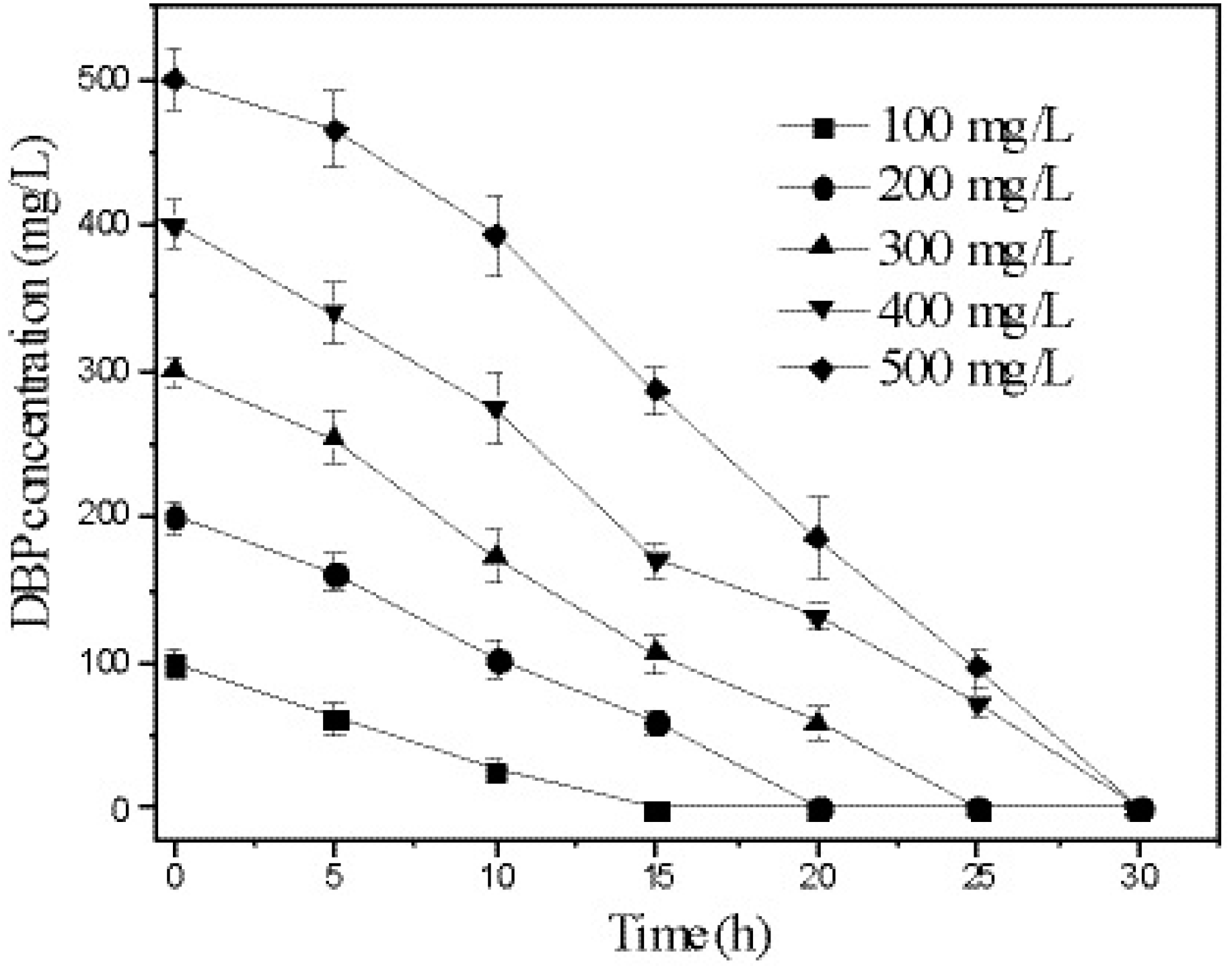
| Initial Concentration (mg/L) | Kinetic Equations | R2 | t1/2 (h) |
|---|---|---|---|
| 100 | lnC = −0.1341 x + 4.6721 | 0.9709 | 5.1 |
| 200 | lnC = −0.0826 x + 5.3931 | 0.9642 | 8.3 |
| 300 | lnC = −0.0825 x + 5.8534 | 0.9623 | 8.4 |
| 400 | lnC = −0.0674 x + 6.1341 | 0.9610 | 10.3 |
| 500 | lnC = −0.0644 x + 6.4367 | 0.9109 | 10.8 |
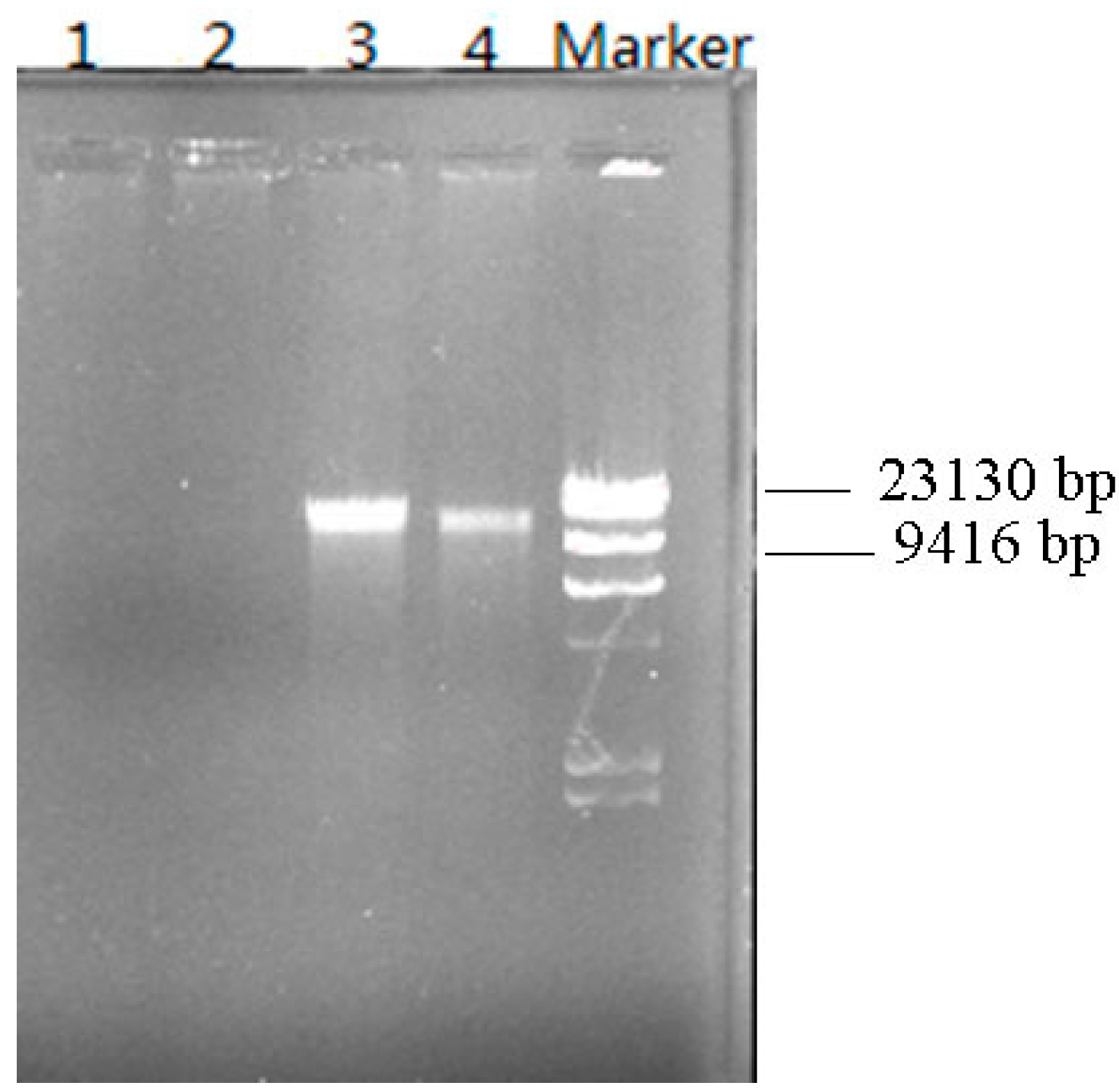
3.6. Plasmid Elimination Experiment
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chi, J.; Gao, J. Effects of Potamogeton crispus L.-bacteria interactions on the removal of phthalate acid esters from surface water. Chemosphere 2015, 119, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Veeramachaneni, D.N. Subchronic exposure to low concentrations of di-n-butyl phthalate disrupts spermatogenesis in Xenopus. laevis frogs. Toxicol. Sci. 2005, 84, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.Y.; Liu, C.; Liao, C.S.; Chang, B.V. Occurrence and microbial degradation of phthalate esters in Taiwan river sediments. Chemosphere 2002, 49, 1295–1299. [Google Scholar] [CrossRef]
- Wang, J.; Chen, G.C.; Christie, P.; Zhang, M.Y.; Luo, Y.M.; Teng, Y. Occurrence and risk assessment of phthalate esters (PAEs) in vegetables and soils of suburban plastic film greenhouses. Sci. Total. Environ. 2015, 523, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Chen, P.Y.; Liu, L.; Zeng, Y.Q.; Zhou, H.X.; Li, S. Effects of combined exposureto17α-ethynylestradiol and dibutyl phthalate on the growth and reproduction of adult male zebrafish (Danio. rerio). Ecotox. Environ. Safe. 2014, 107, 61–70. [Google Scholar]
- Chen, X.; Xu, S.; Tan, T.; Lee, S.T.; Cheng, S.H.; Lee, F.W.F.; Xu, S.J.L.; Ho, K.C. Toxicity and estrogenic endocrine disrupting activity of phthalates and their mixtures. Int. J. Environ. Res. Public Health 2014, 11, 3156–3168. [Google Scholar] [CrossRef] [PubMed]
- Staples, C.A.; Peterson, D.R.; Parkerton, T.F.; Adams, W.J. The environmental fate of phthalic eaters: A literature review. Chemosphere 1997, 35, 667–749. [Google Scholar] [CrossRef]
- Jin, D.C.; Liang, R.X.; Dai, Q.Y.; Zhang, R.Y.; Wu, X.L.; Chao, W.L. Biodegradation of di-n-butyl phthalate by Rhodococcus. sp. JDC-11 and molecular detection of 3,4-phthalate dioxygenase gene. J. Microbiol. Biotechnol. 2010, 20, 1440–1445. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.L.; Wang, Y.Y.; Liang, R.X.; Dai, Q.Y.; Jin, D.C.; Chao, W.L. Biodegradation of an endocrine-disrupting chemical di-n-butyl phthalate by newly isolated Agrobacterium sp. and the biochemical pathway. Process. Biochem. 2011, 46, 1090–1094. [Google Scholar] [CrossRef]
- Jin, D.C.; Bai, Z.H.; Chang, D.D.; Jin, B.; Wang, P.; Wei, D.B.; Zhuang, G.Q. Biodegradation of di-n-butyl phthalate by an isolated Gordonia. sp. strain QH-11: Genetic identification and degradation kinetics. J. Hazard. Mater. 2012, 221–222, 80–85. [Google Scholar]
- Fang, C.R.; Yao, J.; Zheng, Y.G.; Jiang, C.J.; Hu, L.F.; Wu, Y.Y.; Shen, D.S. Dibutyl phthalate degradation by Enterobacter. sp. T5 isolated from municipal solid waste in landfill bioreactor. Int. Biodeter. Biodegr. 2010, 64, 442–446. [Google Scholar] [CrossRef]
- Xu, X.R.; Li, H.B.; Gu, J.D. Biodegradation of an endocrine disrupting chemical di-n-butyl-phthalate ester by Pseudomonas fluorescens B-1. Int. Biodeterior. Biodegrad. 2005, 55, 9–15. [Google Scholar] [CrossRef]
- Chang, H.K.; Zylstra, G.J. Novel organization of the genes for phthalate degradation from Burkholderia. cepacia DB01. J. Bacteriol. 1998, 180, 6529–6537. [Google Scholar] [PubMed]
- Quan, C.S.; Liu, Q.; Tian, W.J.; Fan, S.D. Biodegradation of an endocrine-disrupting chemical, di-2-ethyhexylphthalate by Bacillus subtilis No.66. Appl. Microbiol. Biotechnol. 2005, 66, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.S.; Chen, L.C.; Chen, B.S.; Lin, S.H. Bioremediation of endocrine disruptor di-n-butyl phthalate ester by Deinococcus. radiodurans and Pseudomonas stutzeri. Chemosphere 2010, 78, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Vamsee-Krishna, C.; Phale, P.S. Bacterial degradation of phthalate isomers and their esters. Indian J. Microbiol. 2008, 48, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Habe, H.; Miyakoshi, M.; Chung, J.; Kasuga, K.; Yoshida, T.; Nojiri, H.; Omori, T. Phthalate catabolic gene cluster is linked to the angular dioxygenase gene in Terrabacter. sp. strain DBF 63. Appl. Microbiol. Biotechnol. 2003, 6, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Eaton, R.W. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 2001, 183, 3689–3703. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, T.; Iwata, M.; Imaoka, T.; Mutoh, M.; Egashira, Y.; Nishiyama, T.; Shin, T.; Fujii, T. A mono-2-ethylhexyl phthalate hydrolase from a Gordonia. sp. that is able to dissimilate di-2-ethylhexyl phthalate. Appl. Environ. Microbiol. 2006, 72, 2394–2399. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liao, X.W.; Yu, F.B.; Wei, Z.B.; Yang, L.Y. Cloning of a dibutyl phthalate hydrolase gene from Acinetobacter. sp. strain M673 and functional analysis of its expression product in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 2483–2491. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Fan, X.; Qiu, Y.J.; Li, C.Y.; Xing, S.; Zheng, Y.T.; Xu, J.H. Newly identified thermostable esterase from Sulfobacillus. acidophilus: properties and performance in phthalate ester degradation. Appl. Environ. Microbiol. 2014, 80, 6870–6878. [Google Scholar] [CrossRef] [PubMed]
- El-Mansi, M.; Anderson, K.J.; Inche, C.A.; Knowles, L.K.; Platt, D.J. Isolation and curing of the Klebsiella. pneumoniae large indigenous plasmid using sodium dodecyl sulphate. Res. Microbiol. 2000, 151, 201–208. [Google Scholar] [CrossRef]
- Wan, N.S.; Gu, J.D.; Yan, Y. Degradation of p-nitrophenol by Achromobacter. xylosoxidans Ns isolated from wetland sediment. Int. Biodeterior. Biodegrad. 2007, 58, 99–105. [Google Scholar] [CrossRef]
- Quan, X.C.; Shi, H.C.; Zhang, Y.M.; Wang, J.L.; Qian, Y. Biodegradation of 2,4-dichlorophenol and phenol in an airlift inner-loop bioreactor immobilized with Achromobacter. sp. Sep. Purif. Technol. 2004, 34, 97–103. [Google Scholar] [CrossRef]
- Xu, C.; Ding, J.H.; Qiu, J.G.; Ma, Y. Biodegradation of acetochlor by a newly isolated Achromobacter. sp. strain D-12. J. Environ. Sci. Health B 2013, 48, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.M.; Zhu, W.J.; Long, H.Z.; Chai, L.Y.; Wang, Q.W. Chromate reduction by resting cells of Achromobacter. sp. Ch-1 under aerobic conditions. Process. Biochem. 2007, 42, 1028–1032. [Google Scholar] [CrossRef]
- Pradeep, S.; Sarath Josh, M.K.; Binod, P.; Sudha Devi, R.; Balachandran, S.; Anderson, R.C.; Benjamin, S. Achromobacter. denitrificans strain SP1 efficiently remediates di (2-ethylhexyl) phthalate. Ecotoxicol. Environ. Saf. 2015, 112, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.C.; Kong, X.; Cui, B.J.; Bai, Z.H.; Zhang, H.X. Biodegradation of di-n-butyl phthalate by a newly isolated halotolerant Sphingobium. sp. Int. J. Mol. Sci. 2013, 14, 24046–24054. [Google Scholar] [CrossRef] [PubMed]
- Vedler, E.; Kõiv, V.; Heinaru, A. Analysis of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pEST4011 of Achromobacter. xylosoxidans subsp. denitrificans strain EST4002. Gene 2000, 255, 281–288. [Google Scholar] [PubMed]
- Strnad, H.; Ridl, J.; Paces, J.; Kolar, M.; Vlcek, C.; Paces, V. Complete genome sequence of the haloaromatic acid-degrading bacterium Achromobacter. xylosoxidans A8. J. Bacteriol. 2011, 193, 791–792. [Google Scholar] [CrossRef] [PubMed]
- Nomura, Y.; Nakagawa, M.; Ogawa, N.; Harashima, S.; Oshima, Y. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J. Ferment. Bioeng. 1992, 74, 333–344. [Google Scholar] [CrossRef]
- Karpagam, S.; Lalithakumari, D. Plasmid mediated degradation of o-and p-phthalate by Pseudomonas fluorescens. W. J. Microbiol. Biotechnol. 1999, 15, 565–569. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, D.; Kong, X.; Li, Y.; Bai, Z.; Zhuang, G.; Zhuang, X.; Deng, Y. Biodegradation of di-n-Butyl Phthalate by Achromobacter sp. Isolated from Rural Domestic Wastewater. Int. J. Environ. Res. Public Health 2015, 12, 13510-13522. https://doi.org/10.3390/ijerph121013510
Jin D, Kong X, Li Y, Bai Z, Zhuang G, Zhuang X, Deng Y. Biodegradation of di-n-Butyl Phthalate by Achromobacter sp. Isolated from Rural Domestic Wastewater. International Journal of Environmental Research and Public Health. 2015; 12(10):13510-13522. https://doi.org/10.3390/ijerph121013510
Chicago/Turabian StyleJin, Decai, Xiao Kong, Yujie Li, Zhihui Bai, Guoqiang Zhuang, Xuliang Zhuang, and Ye Deng. 2015. "Biodegradation of di-n-Butyl Phthalate by Achromobacter sp. Isolated from Rural Domestic Wastewater" International Journal of Environmental Research and Public Health 12, no. 10: 13510-13522. https://doi.org/10.3390/ijerph121013510







