Infectious Risk Assessment of Unsafe Handling Practices and Management of Clinical Solid Waste
Abstract
:1. Introduction
2. Experimental Section
2.1. Clinical Solid Waste Collection and Preparation
2.2. Identification of the Bacteria in Clinical Solid Waste Using the Conventional Method
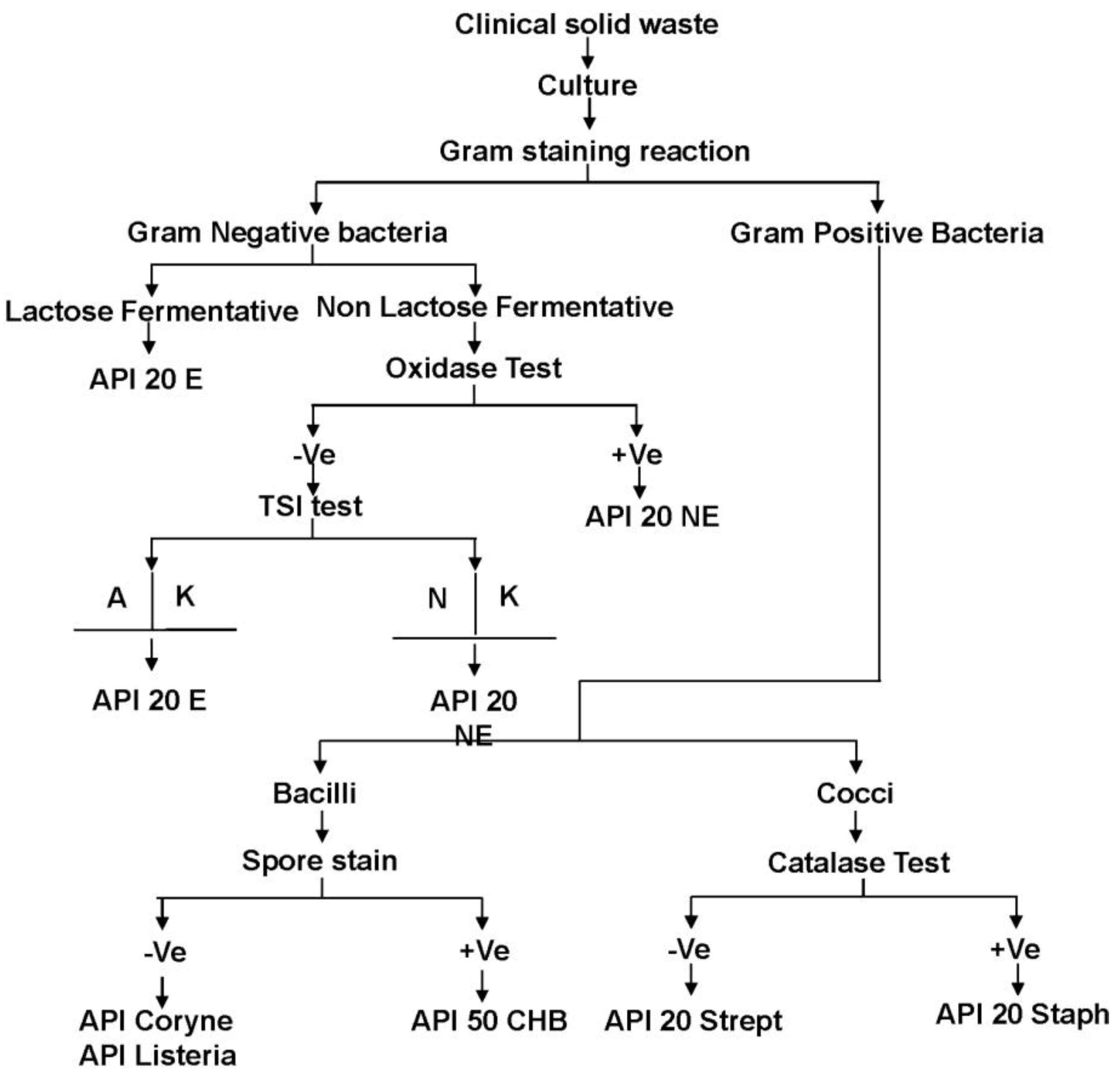
2.3. Identification of the Bacteria in Clinical Sharp Waste Using Genomic DNA Profiling
3. Results and Discussion
| No. | Type of clinical solid waste | Microorganisms |
|---|---|---|
| 1 | General waste | Escherichia coli |
| Proteus mirabilis | ||
| Pseudomonas aeruginosa | ||
| Staphylococcus aureus | ||
| 2 | Dental solid waste | Streptococcus mutans |
| Staphylococcus epidermidis | ||
| Salmonella spp. | ||
| Enterococcus faecalis | ||
| Klebsiella pneumoniae | ||
| Serratia liquefaciens | ||
| Escherichia coli | ||
| Staphylococcus aureus | ||
| 3 | Microbiological lab waste | Streptococcus pyogenes |
| Streptococcus mutans | ||
| Escherichia coli | ||
| Klebsiella pneumoniae | ||
| Serratia liquefaciens | ||
| Serratia marcescens | ||
| Proteus mirabilis | ||
| Enterococcus faecalis | ||
| Staphylococcus aureus | ||
| Staphylococcus epidermidis | ||
| Staphylococcus lentus | ||
| Pseudomonas aeruginosa | ||
| Lysinibacillus sphaericus | ||
| Pseudomonas aeruginosa | ||
| Acinetobacter lwoffii | ||
| Acinetobacter baumannii | ||
| 4 | Clinical solid waste from ICU unit | Acinetobacter baumannii |
| Klebsiella pneumoniae | ||
| Proteus mirabilis | ||
| Staphylococcus aureus | ||
| Pseudomonas aeruginosa | ||
| Escherichia coli | ||
| Serratia marcescens | ||
| 5 | Clinical solid waste from Dermatology unit | Streptococcus pyogenes |
| Staphylococcus aureus | ||
| Staphylococcus epidermidis | ||
| Pseudomonas aeruginosa | ||
| 6 | Clinical solid waste from Isolation wards | Streptococcus pyogenes |
| Staphylococcus aureus | ||
| Staphylococcus epidermidis | ||
| Enterococcus faecalis | ||
| Escherichia coli | ||
| Klebsiella pneumoniae | ||
| Enterococcus faecalis | ||
| Proteus mirabilis | ||
| Pseudomonas aeruginosa | ||
| Lysinibacillus sphaericus | ||
| 7 | Clinical solid waste from Obstetrical & Gynecology ward | Escherichia coli |
| Pseudomonas aeruginosa | ||
| Proteus mirabilis | ||
| Streptococcus agalactiae |
| Exhibiting ≥99% 16s rDNA sequence homology with the blast results | Exhibiting <99% 16s rDNA sequence homology with the blast results similarity | ||
|---|---|---|---|
| Bacterial Strain | Accession ID | Bacterial Strain | Accession ID |
| Escherichia coli strain D i14 | CP002212.1 | Escherichia coli strain 114 | JN180963.1 |
| Escherichia coli strain KO11FL | CP002970.1 | Escherichia coli strain APEC O1 | CP000468.1 |
| Escherichia coli strain UM146 | CP002167.1 | Escherichia coli strain BW2952 | CP001396.1 |
| Klebsiella pneumoniae strain AGR/IICT/1 | JQ973896.1 | Escherichia coli strain IHE3034 | CP001969.1 |
| Klebsiella pneumoniae strain mcp11d | EF419182.1 | Klebsiella pneumoniae strain ELA-21o | FJ195012.1 |
| Proteus mirabilis strain HH134 | HQ407314.1 | Klebsiella pneumoniae strain T89 16S | HQ407264.1 |
| Proteus mirabilis strain HH139 | HQ407311.1 | Lysinibacillus sphaericus strain NBIGP 6 | JF304288.1 |
| Serratia marcescens strain 21-3 | JF429937.1 | Lysinibacillus sphaericus strain SP19_LP11 | JQ289050.1 |
| Serratia marcescens strain CTC639-K12 | JQ917918.1 | ||
| Staphylococcus aureus strain HO 5096 0412 | HE681097.1 | ||
| Staphylococcus aureus strain NBRC 102140 | AB681714.1 | ||
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Bdour, A.; Altrabsheh, B.; Hadadin, N.; Al-Shareif, M. Assessment of medical wastes management practice: A case study of the northern part of Jordan. Waste Management 2007, 27, 746–759. [Google Scholar] [CrossRef]
- Hossain, M.S.; Santhanam, A.; Nik Norulaini, N.A.; Mohd Omar, A.K. Clinical solid waste management practices and its impact on human health and environment—A review. Waste Management 2011, 31, 754–766. [Google Scholar]
- Brent, A.C.; Rogers, D.E.C.; Ramabitsa-Siimane, T.S.M.; Rohwer, M.B. Application of the analytical hierarchy process to establish health care waste management systems that minimise infection risks in developing countries. Eur. J. Oper. Res. 2007, 181, 403–424. [Google Scholar]
- Siegel, J.D.; Rhinehart, E.; Jackson, M.; Chiarello, L. 2007 Guideline for isolation precautions: Preventing transmission of infectious agents in health care settings. Amer. J. Infect. Control 2007, 35, S65–S164. [Google Scholar] [CrossRef]
- Abd El-Salam, M.M. Hospital waste management in El-Beheira Governorate, Egypt. J. Environ. Manage. 2010, 91, 618–629. [Google Scholar] [CrossRef]
- Al-Khatib, I.A.; Sato, C. Solid health care waste management status at health care centers in the West Bank—Palestinian Territory. Waste Management 2009, 29, 2398–2403. [Google Scholar] [CrossRef]
- Nemathaga, F.; Maringa, S.; Chimuka, L. Hospital solid waste management practices in Limpopo Province, South Africa: A case study of two hospitals. Waste Management 2008, 28, 1236–1245. [Google Scholar] [CrossRef]
- Singh, R.P.; Singh, P.; Araujo, A.S.F.; Hakimi Ibrahim, M.; Sulaiman, O. Management of urban solid waste: Vermicomposting a sustainable option. Resour. Conserv. Recycl. 2011, 55, 719–729. [Google Scholar] [CrossRef]
- Coker, A.; Sangodoyin, A.; Sridhar, M.; Booth, C.; Olomolaiye, P.; Hammond, F. Medical waste management in Ibadan, Nigeria: Obstacles and prospects. Waste Management 2009, 29, 804–811. [Google Scholar]
- Al-Sohaibani, M.O.; Al-Sheikh, E.H.; Al-Ballal, S.J.; Mirghani, M.A.M.; Ramia, S. Occupational risk of hepatitis B and C infections in Saudi medical staff. J. Hosp. Infect. 1995, 31, 143–147. [Google Scholar]
- Franka, E.; El-Zoka, A.H.; Hussein, A.H.; Elbakosh, M.M.; Arafa, A.K.; Ghenghesh, K.S. Hepatitis B virus and hepatitis C virus in medical waste handlers in Tripoli, Libya. J. Hosp. Infect. 2009, 72, 258–261. [Google Scholar] [CrossRef]
- Gunson, R.N.; Shouval, D.; Roggendorf, M.; Zaaijer, H.; Nicholas, H.; Holzmann, H.; de Schryver, A.; Reynders, D.; Connell, J.; Gerlich, W.H.; Marinho, R.T.; Tsantoulas, D.; Rigopoulou, E.; Rosenheim, M.; Valla, D.; Puro, V.; Struwe, J.; Tedder, R.; Aitken, C.; Alter, M.; Schalm, S.W.; Carman, W.F. Hepatitis B virus (HBV) and hepatitis C virus (HCV) infections in health care workers (HCWs): Guidelines for prevention of transmission of HBV and HCV from HCW to patients. J. Clin. Virol. 2003, 27, 213–230. [Google Scholar] [CrossRef]
- WHO, Safe Health-Care Waste Management: Policy Paper by the World Health Organisation; Department of Protection of the Human Environment, Water, Sanitation and Health: Geneva, Switzerland, 2005.
- Blenkharn, J.I. Safe disposal and effective destruction of clinical wastes. J. Hosp. Infect. 2005, 60, 295–297. [Google Scholar] [CrossRef]
- Borg, M.A. Clinical waste disposal—Getting the facts right. J. Hosp. Infect. 2007, 65, 178–180. [Google Scholar]
- Alagöz, A.Z.; Kocasoy, G. Determination of the best appropriate management methods for the health-care wastes in İstanbul. Waste Management 2008, 28, 1227–1235. [Google Scholar] [CrossRef]
- Saini, S.; Das, K.B.; Kapil, A.; Nagargan, S.; Sarma, R.K. The Study of bacterial flora of different types in hospital waste: Evaluation of waste treatment at aims hospital, New Delhi. Bacter. Flora Hospit. Waste 2004, 35, 986–989. [Google Scholar]
- Pruss, A.; Giroult, E.; Rushbrook, P. Safe Management of Wastes from Healthcare Activities; World Health Organization: Geneva, Switzerland, 1999. [Google Scholar]
- Park, H.; Lee, H.; Kim, M.; Lee, J.; Seong, S.-Y.; Ko, G. Detection and hazard assessment of pathogenic microorganisms in medical waste. J. Environ. Sci. Health Part A 2009, 44, 995–1003. [Google Scholar] [CrossRef]
- Ministry of Health Guideline on the Handling and Management of Clinical Waste in Malaysia, 3rd ed; Department of Environment, Federal Government Administrative Centre: Putrajaya, Malaysia, 2009.
- Lennette, H.E.; Spaulding, H.E.; Ruant, J.P. Manual of Clinical Microbiology, 5th ed; American society for microbiology: Washington, DC, USA, 1991. [Google Scholar]
- Alagöz, A.Z.; Kocasoy, G. Determination of the best appropriate management methods for the health-care wastes in İstanbul. Waste Management 2008, 28, 1227–1235. [Google Scholar] [CrossRef]
- Vieira, C.D.; de Carvalho, M.A.R.; de Menezes Cussiol, N.A.; Alvarez-Leite, M.E.; dos Santos, S.G.; da Fonseca Gomes, R.M.; Silva, M.X.; Nicoli, J.R.; de Macêdo Farias, L. Count, identification and antimicrobial susceptibility of bacteria recovered from dental solid waste in Brazil. Waste Management 2011, 31, 1327–1332. [Google Scholar] [CrossRef]
- Høiby, N. Isolation and treatment of cystic fibrosis patients with lung infections caused by Pseudomonas (Burkholderia) cepacia and multiresistant Pseudomonas aeruginosa. Neth. J. Med. 1995, 46, 280–287. [Google Scholar] [CrossRef]
- Zembrzuska-Sadkowska, E.; Sneum, M.; Ojeniyi, B.; Heiden, L.; Høiby, N. Epidemiology of Pseudomonas aeruginosa infection and the role of contamination of the environment in the Danish Cystic Fibrosis Centre. J. Hosp. Infect. 1995, 29, 1–7. [Google Scholar] [CrossRef]
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar]
- Renois, F.; Jacques, J.; Guillard, T.; Moret, H.; Pluot, M.; Andreoletti, L.; de Champs, C. Preliminary investigation of a mice model of Klebsiella pneumoniae subsp. ozaenae induced pneumonia. Microbes Infect. 2011, 13, 1045–1051. [Google Scholar]
- Coker, A.O.; Akanmu, A.M.; Sridhar, M.K.C.; Aladenola, I.O. Management of Pollution from an Hospital Incinerator in Nigeria; WIT Press: Southampton, UK, 2000. [Google Scholar]
- Tang, Y.-W.; Stratton, C.W. Staphylococcus aureus: An old pathogen with new weapons. Clin. Lab. Med. 2010, 30, 179–208. [Google Scholar] [CrossRef]
- Arciola, C.R.; Campoccia, D.; Baldassarri, L.; Pirini, V.; Huebner, J.; Montanaro, L. The role of Enterococcus faecalis in orthopaedic peri-implant infections demonstrated by automated ribotyping and cluster analysis. Biomaterials 2007, 28, 3987–3995. [Google Scholar] [CrossRef]
- Gómez-Gil, R.; Romero-Gómez, M.P.; García-Arias, A.; Ubeda, M.G.; Busselo, M.S.; Cisterna, R.; Gutiérrez-Altés, A.; Mingorance, J. Nosocomial outbreak of linezolid-resistant Enterococcus faecalis infection in a tertiary care hospital. Diagn. Microbiol. Infect. Dis. 2009, 65, 175–179. [Google Scholar] [CrossRef]
- Bremer, A.A.; Darouiche, R.O. Ventriculoperitoneal shunt infection due to Serratia marcescens. J. Infect. 2005, 50, 138–141. [Google Scholar] [CrossRef]
- García, D.C.; Catalano, M.; Piñeiro, S.; Woloj, M.; Kaufman, S.; Sordelli, D.O. The emergence of resistance to amikacin in Serratia marcescens isolates from patients with nosocomial infection. Int. J. Antimicrobial Agents 1996, 7, 203–210. [Google Scholar]
- Giamarellou, H.; Antoniadou, A.; Kanellakopoulou, K. Acinetobacter baumannii: A universal threat to public health? Int. J. Antimicrobial Agents 2008, 32, 106–119. [Google Scholar]
- Marques, M.B.; Waites, K.B.; Mangino, J.E.; Hines, B.B.; Moser, S.A. Genotypic investigation of multidrug-resistant Acinetobacter baumannii infections in a medical intensive care unit. J. Hosp. Infect. 1997, 37, 125–135. [Google Scholar] [CrossRef]
- Ahmed, I.; Yokota, A.; Yamazoe, A.; Fujiwara, T. Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int. J. Syst. Evol. Microbial. 2007, 57, 1117–1125. [Google Scholar] [CrossRef]
- Avasthi, T.S.; Kumar, N.; Baddam, R.; Hussain, A.; Nandanwar, N.; Jadhav, S.; Ahmed, N. Genome of multidrug-resistant uropathogenic Escherichia coli strain NA114 from India. J. Bacteriol. 2011, 93, 4272–4273. [Google Scholar]
- Sawalem, M.; Selic, E.; Herbell, J.D. Hospital waste management in Libya: A case study. Waste Management 2009, 29, 1370–1375. [Google Scholar] [CrossRef]
- Matsui, M.; Kashima, Y.; Kawano, M.; Matsuda, M.; Ambe, K.; Wakimoto, T.; Doi, R. Dioxin-like potencies and extractable organohalogens (EOX) in medical, municipal and domestic waste incinerator ashes in Japan. Chemosphere 2003, 53, 971–980. [Google Scholar] [CrossRef]
- Tsakona, M.; Anagnostopoulou, E.; Gidarakos, E. Hospital waste management and toxicity evaluation: A case study. Waste Management 2007, 27, 912–920. [Google Scholar] [CrossRef]
- Lee, B.-K.; Ellenbecker, M.J.; Moure-Ersaso, R. Alternatives for treatment and disposal cost reduction of regulated medical wastes. Waste Management 2004, 24, 143–151. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hossain, M.S.; Rahman, N.N.N.A.; Balakrishnan, V.; Puvanesuaran, V.R.; Sarker, M.Z.I.; Kadir, M.O.A. Infectious Risk Assessment of Unsafe Handling Practices and Management of Clinical Solid Waste. Int. J. Environ. Res. Public Health 2013, 10, 556-567. https://doi.org/10.3390/ijerph10020556
Hossain MS, Rahman NNNA, Balakrishnan V, Puvanesuaran VR, Sarker MZI, Kadir MOA. Infectious Risk Assessment of Unsafe Handling Practices and Management of Clinical Solid Waste. International Journal of Environmental Research and Public Health. 2013; 10(2):556-567. https://doi.org/10.3390/ijerph10020556
Chicago/Turabian StyleHossain, Md. Sohrab, Nik Norulaini Nik Ab Rahman, Venugopal Balakrishnan, Vignesh R. Puvanesuaran, Md. Zaidul Islam Sarker, and Mohd Omar Ab Kadir. 2013. "Infectious Risk Assessment of Unsafe Handling Practices and Management of Clinical Solid Waste" International Journal of Environmental Research and Public Health 10, no. 2: 556-567. https://doi.org/10.3390/ijerph10020556
APA StyleHossain, M. S., Rahman, N. N. N. A., Balakrishnan, V., Puvanesuaran, V. R., Sarker, M. Z. I., & Kadir, M. O. A. (2013). Infectious Risk Assessment of Unsafe Handling Practices and Management of Clinical Solid Waste. International Journal of Environmental Research and Public Health, 10(2), 556-567. https://doi.org/10.3390/ijerph10020556





