Genetic Approach for the Fast Discovery of Phenazine Producing Bacteria
Abstract
:1. Introduction
2. Results and Discussion
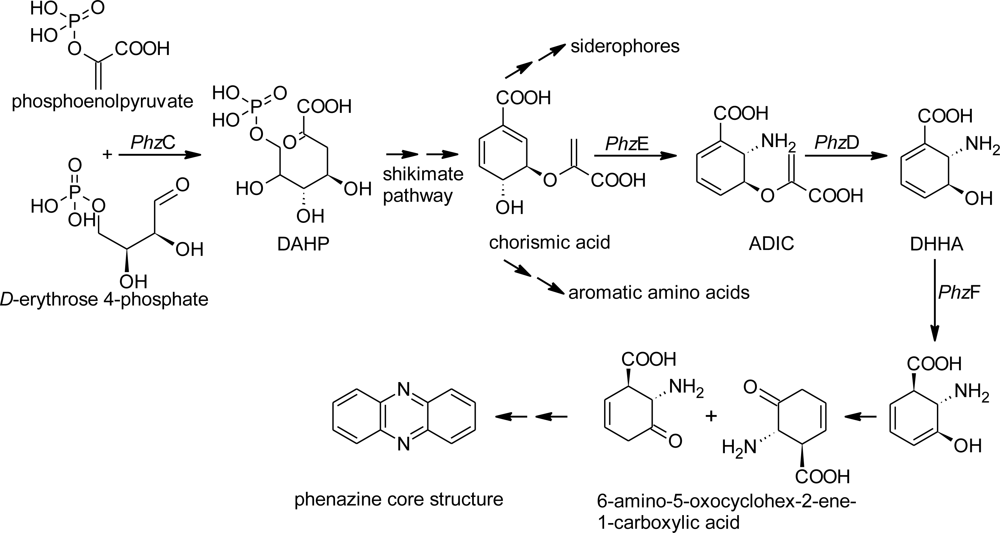
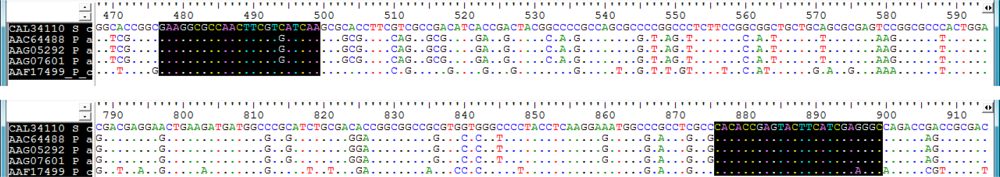
2.1. Design of Oligonucleotides to Search for phzE Phenazine Gene Fragments
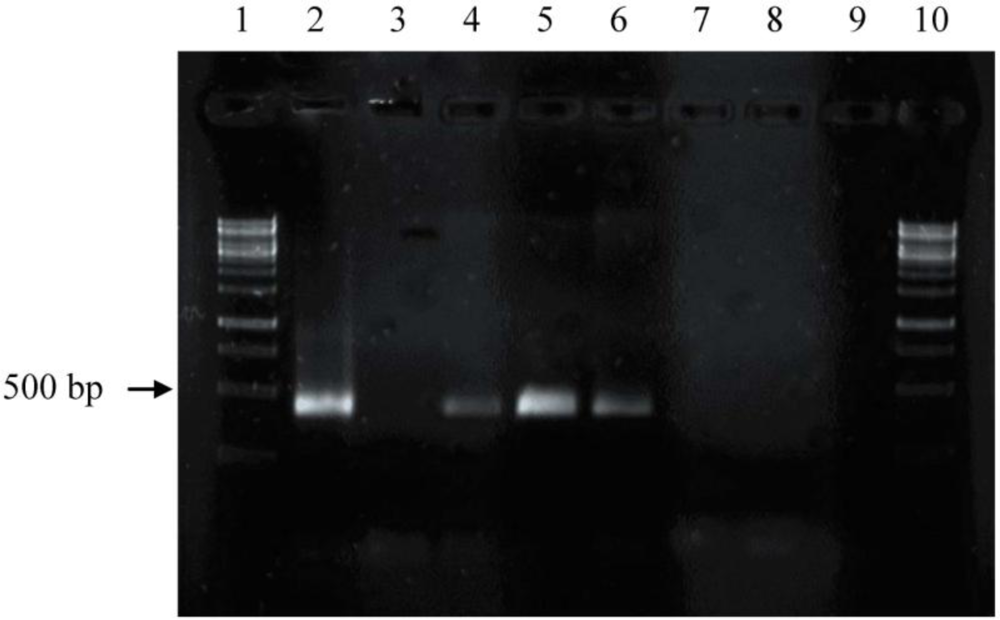
2.2. Screening for phzE Gene Fragments with the Constructed Primers
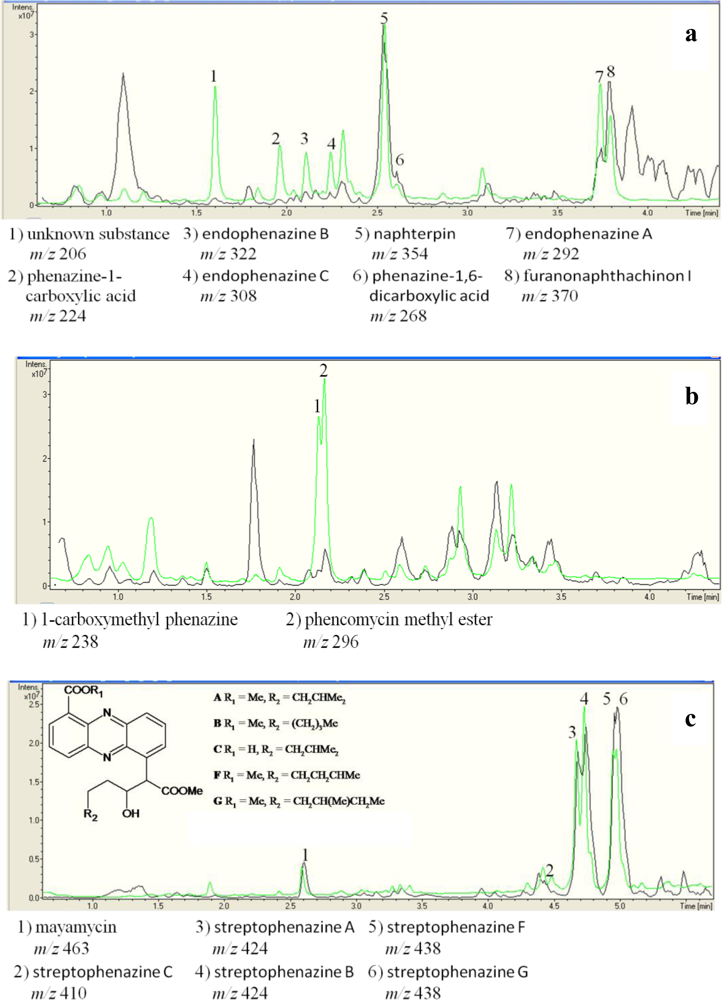
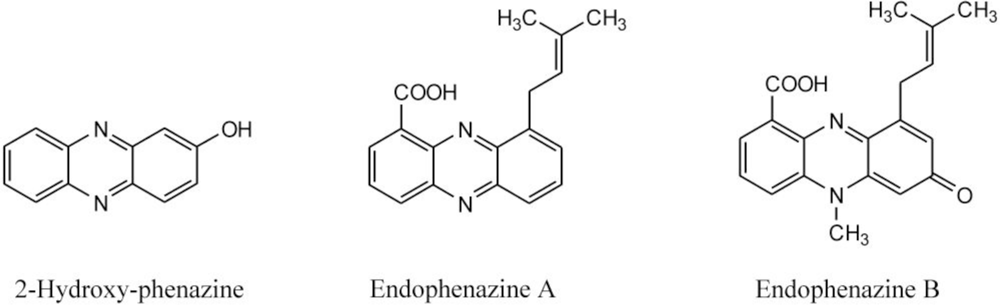
2.3. Detection of Phenazines in the phzE Positive Strains
3. Experimental Section
3.1. Bacterial Strains and Their Phylogenetic Affiliation
3.2. Design of Oligonucleotides for Molecular Detection of phzE Phenazine Gene Fragments
3.3. Amplification and Identification of the Phenazine Gene Fragments
3.4. Cultivation of phzE Strains
3.4.1. Cultivation of phzE Positive Strain
3.4.2. Cultivation of phzE Negative Strains
3.5. Culture Extracts of phzE Positive and Negative Strains
3.6. Chemical Analysis of phzE Positive and Negative Strains
3.7. Nucleotide Sequence Accession Numbers
4. Conclusions
Acknowledgments
- Samples Availability: Available from the authors.
References
- Blunt, JW; Copp, BR; Munro, MHG; Northcote, PT; Prinsep, MR. Marine natural products. Nat Prod Rep 2011, 28, 196–268. [Google Scholar]
- Newman, DJ; Cragg, GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod 2007, 70, 461–447. [Google Scholar]
- Butler, MS. Natural products to drugs: Natural product-derived compounds in clinical trials. Nat Prod Rep 2008, 25, 475–516. [Google Scholar]
- Donadio, S; Monciardini, P; Sosio, M. Polyketide synthases and nonribosomal peptide synthetases: The emerging view from bacterial genomics. Nat Prod Rep 2007, 24, 1073–1109. [Google Scholar]
- Willey, JM; van der Donk, WA. Lantibiotics: Peptides of diverse structure and function. Annu Rev Microbiol 2007, 61, 477–501. [Google Scholar]
- Arbiser, JL; Moschella, SL. Clofazimine: A review of its medical uses and mechanisms of action. J Am Acad Dermatol 1995, 32, 241–247. [Google Scholar]
- McDonald, M; Mavrodi, DV; Thomashow, LS; Floss, HG. Phenazine biosynthesis in Pseudomonas fluorescens: Branchpoint from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. J Am Chem Soc 2001, 123, 9459–9460. [Google Scholar]
- Pierson, LS, III; Gaffney, T; Lam, S; Gong, F. Molecular analysis of genes encoding phenazine biosynthesis in the biological control bacterium Pseudomonas aureofaciens 30–84. FEMS Microbiol Lett 1995, 134, 299–307. [Google Scholar]
- Pusecker, K; Laatsch, H; Helmke, E; Weyland, H. Dihydrophencomycin methyl ester, a new phenazine derivative from a marine Streptomycete. J Antibiot 1997, 50, 479–483. [Google Scholar]
- Van Niekerk, S; Huygens, F; van Rensburg, CEJ. A time-kill study to evaluate the in vitro activity of clofazimine in combination with cefotaxime against a penicillin- and cefotaxime-resistant strain of Streptococcus pneumoniae. J Antimicrob Chemother 1997, 40, 602–604. [Google Scholar]
- Van Rensburg, CEJ; Joone, GK; O’Sullivan, JF; Anderson, R. Antimicrobial activities of clofazimine and B669 are mediated by lysophospholipids. Antimicrob Agents Chemother 1992, 36, 2729–2735. [Google Scholar]
- Mavrodi, DV; Blankenfeldt, W; Thomashow, LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol 2006, 44, 417–445. [Google Scholar]
- Reddy, VM; O’Sullivan, JF; Gangadharam, PR. Antimycobacterial activities of riminophenazines. J Antimicrob Chemother 1999, 43, 615–623. [Google Scholar]
- Spicer, JA; Gamage, SA; Rewcastle, GW; Finlay, GJ; Bridewell, DJ; Baguley, BC; Denny, WA. Bis(phenazine-1-carboxamides): Structure-activity relationships for a new class of dual topoisomerase I/II-directed anticancer drugs. J Med Chem 2000, 43, 1350–1358. [Google Scholar]
- Haagen, Y; Glück, K; Fay, K; Kammerer, B; Gust, B; Heide, L. A gene cluster for prenylated naphthoquinone and prenylated phenazine biosynthesis in Streptomyces cinnamonensis DSM 1042. Chembiochem 2006, 7, 2016–2027. [Google Scholar]
- Mavrodi, DV; Ksenzenko, VN; Bonsall, RF; Cook, RJ; Boronin, AM; Thomashow, LS. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J Bacteriol 1998, 180, 2541–2548. [Google Scholar]
- Mentel, M; Ahuja, EG; Mavrodi, DV; Breinbauer, R; Thomashow, LS; Blankenfeldt, W. Of two make one: The biosynthesis of phenazines. Chembiochem 2009, 10, 2295–2304. [Google Scholar]
- McDonald, M; Mavrodi, DV; Thomashow, LS; Floss, HG. Phenazine biosynthesis in Pseudomonas fluorescens: Branchpoint from the primary shikimate biosynthetic pathway and role of phenazine-1,6-dicarboxylic acid. J Am Chem Soc 2001, 123, 9459–9460. [Google Scholar]
- Pierson, LS, III; Pierson, EA. Metabolism and function of phenazines in bacteria: Impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotech 2010, 86, 1659–1670. [Google Scholar]
- Calhoun, DH; Carson, M; Jensen, RA. The branch point metabolite for pyocyanine biosynthesis in Pseudomonas aeruginosa. J Gen Microbiol 1972, 72, 581–583. [Google Scholar]
- Mavrodi, DV; Peever, TL; Mavrodi, OV; Parejko, JA; Raaijmakers, JM; lemanceau, P; Mazurier, S; Heide, L; Blankenfeldt, W; Weller, DM; et al. Diversity and evolution of the phenazine biosynthesis pathway. Appl Environ Microbiol 2010, 76, 866–879. [Google Scholar]
- Fiedler, HP; Bruntner, C; Bull, AT; Ward, AC; Goodfellow, M; Potterat, O; Puder, C; Mihm, G. Marine actinomycetes as a source of novel secondary metabolites. Anton Leeuwenhoek 2005, 87, 37–42. [Google Scholar]
- Jensen, PR; Mincer, TJ; Williams, PG; Fenical, W. Marine actinomycete diversity and natural product discovery. Anton Leeuwenhoek 2005, 87, 43–48. [Google Scholar]
- Salomon, CE; Magarvey, NA; Sherman, DH. Merging the potential of microbial genetics with biological and chemical diversity: An even brighter future for marine natural product drug discovery. Nat Prod Rep 2004, 21, 105–121. [Google Scholar]
- Williams, PG. Panning for chemical gold: Marine bacteria as a source of new therapeutics. Trends Biotech 2009, 27, 45–52. [Google Scholar]
- Gulder, TAM; Moore, BS. Chasing the treasures of the sea—Bacterial marine natural products. Curr Opin Microbiol 2009, 12, 252–260. [Google Scholar]
- Metsä-Ketelä, M; Salo, V; Halo, L; Hautala, A; Hakala, J; Mäntsälä, P; Ylihonko, K. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol Lett 1999, 180, 1–6. [Google Scholar]
- Piel, J. A polyketide synthase-peptide synthetases gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA 2002, 99, 14002–14007. [Google Scholar]
- Laursen, JB; de Visser, PC; Nielsen, HK; Jensen, KJ; Nielsen, J. Solid-phase synthesis of new saphenamycin analogues with antimicrobial activity. Bioorg Med Chem Lett 2002, 12, 171–175. [Google Scholar]
- Price-Whelan, A; Dietrich, LE; Newman, DK. Rethinking ‘secondary’ metabolism: Physiological roles for phenazine antibiotics. Nat Chem Biol 2006, 2, 71–78. [Google Scholar]
- Thomashow, LS; Weller, DM; Bonsall, RF; Pierson, LS. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol 1990, 56, 908–912. [Google Scholar]
- Wilson, R; Sykes, DA; Watson, D; Rutman, A; Taylor, GW; Cole, PJ. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect Immun 1988, 56, 2515–2517. [Google Scholar]
- Angell, S; Bench, BJ; Williams, H; Watanabe, CMH. Pyocyanin isolated from a marine microbial population: Synergistic production between two distinct bacterial species and mode of action. Chem Biol 2006, 13, 1349–1359. [Google Scholar]
- Isnansetyo, A; Kamei, Y. Bioactive substances produced by marine isolates of Pseudomonas. J Ind Microbiol Biotech 2009, 36, 1239–1248. [Google Scholar]
- Jayatilake, GS; Thornton, MP; Leonard, AC; Grimwade, JE; Baker, BJ. Metabolites from an Antarctic sponge-associated bacterium, Pseudomonas aeruginosa. J Nat Prod 1996, 59, 293–296. [Google Scholar]
- Imamura, N; Nishijima, M; Takadera, T; Adachi, K; Sakai, M; Sano, H. New anticancer antibiotics pelagiomicins, produced by a new marine bacterium Pelagiobacter variabilis. J Antibiot 1997, 50, 8–12. [Google Scholar]
- Choi, EJ; Kwon, HC; Ham, J; Yang, HO. 6-Hydroxymethyl-1-phenazine-carboxamide and 1,6-phenazinedimethanol from a marine bacterium, Brevibacterium sp. KMD 003, associated with marine purple vase sponge. J Antibiot 2009, 62, 621–624. [Google Scholar]
- Li, D; Wang, F; Xiao, X; Zeng, X; Gu, QQ; Zhu, W. A new cytotoxic phenazine derivative from a deep sea bacterium Bacillus sp. Arch Pharm Res 2007, 30, 552–555. [Google Scholar]
- Mitova, MI; Lang, G; Wiese, J; Imhoff, JF. Subinhibitory concentrations of antibiotics induce phenazine production in a marine Streptomyces sp. J Nat Prod 2008, 71, 824–827. [Google Scholar]
- Turner, JM; Messenger, AJ. Occurrence, biochemistry and physiology of phenazine pigment production. Adv Microb Physiol 1986, 27, 211–275. [Google Scholar]
- Brisbane, PG; Janik, LJ; Tate, ME; Warren, RF. Revised structure for the phenazine antibiotic from Pseudomonas fluorescens 2–79 (NRRL B-15132). Antimicrob Agents Chemother 1987, 31, 1967–1971. [Google Scholar]
- Levitch, ME; Rietz, P. The isolation and characterization of 2-hydroxyphenazine from Pseudomonas aureofaciens. Biochemistry 1966, 5, 689–692. [Google Scholar]
- Kanner, D; Gerber, NN; Bartha, R. Pattern of phenazine pigment production by a strain of Pseudomonas aeruginosa. J Bacteriol 1978, 134, 690–692. [Google Scholar]
- Kunigami, T; Shin-Ya, K; Furihata, K; Furihata, K; Hayakawa, Y; Seto, H. A novel neuronal cell protecting substance, aestivophoenin C, produced by Streptomyces purpeofuscus. J Antibiot 1998, 51, 880–882. [Google Scholar]
- Nakano, H; Yoshida, M; Shirahata, K; Ishii, S; Arai, Y; Morimoto, M; Tomita, F. Senacarcin A, a new antitumor antibiotic produced by Streptomyces endus subsp aureus. J Antibiot 1982, 35, 760–762. [Google Scholar]
- Schneemann, I; Nagel, K; Kajahn, I; Labes, A; Wiese, J; Imhoff, JF. Comprehensive investigations of marine Actinobacteria associated with the sponge Halichondria panicea. Appl Environ Microbiol 2010, 76, 3702–3714. [Google Scholar]
- Wiese, J; Thiel, V; Nagel, K; Staufenberger, T; Imhoff, JF. Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar Biotech 2009, 11, 287–300. [Google Scholar]
- Peix, A; Valverde, A; Rivas, R; Igual, JM; Ramírez-Bahena, MH; Mateos, PF; Santa-Regina, I; Rodríguez-Barrueco, C; Martínez-Molina, E; Velázquez, E. Reclassification of Pseudomonas aurantiaca as a synonym of Pseudomonas chlororaphis and proposal of three subspecies, P. chlororaphis subsp. chlororaphis subsp. nov., P. chlororaphis subsp. aureofaciens subsp. nov., comb. nov. and P. chlororaphis subsp. aurantiaca subsp. nov., comb. nov. Int J Syst Evol Microbiol 2007, 57, 1286–1290. [Google Scholar]
- Thiel, V; Neulinger, SC; Staufenberger, T; Schmaljohann, R; Imhoff, JF. Spatial distribution of sponge-associated bacteria in the Mediterranean sponge Tethya aurantium. FEMS Microbiol Ecol 2007, 59, 47–63. [Google Scholar]
- Altschul, SF; Gish, W; Miller, W; Myers, EW; Lipman, DJ. Basic Local alignment search tool. J Mol Biol 1990, 215, 403–410. [Google Scholar]
- Cole, JR; Wang, Q; Cardenas, E; Fish, J; Chai, B; Farris, RJ; Klam-Syed-Mohideen, AS; McCarell, DM; Marsh, T; Garrity, GM; et al. The Ribosoaml Database Project: Improved alignments and new tools for rRNA analysis. Nucl Acids Res 2009, 37, D141–D145. [Google Scholar]
- Thompson, JD; Gibson, TJ; Plewniak, F; Jeanmougin, F; Higgins, DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality tools. Nucl Acid Res 1997, 25, 4876–4882. [Google Scholar]
- Ashenafi, M; Carrington, R; Collins, AC; Byrnes, WM. The fused TrpEG from Streptomyces venezuelae is an anthranilate synthase, not an 2-amino-4-deoxyisochorismate (ADIC) synthase. Ethn Dis 2008, 18(Suppl. 2), S2–S9–13. [Google Scholar]
- Weisburg, WG; Barns, SM; Pelletier, DA; Lane, DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 1991, 173, 697–703. [Google Scholar]
- Muyzer, G; de Waal, EC; Uitterlinden, AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. J Mol Biol 1993, 59, 695–700. [Google Scholar]
- King, EO; Ward, M; Raney, DE. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med 1954, 44, 301–307. [Google Scholar]
- Laatsch, H. Antibase 2007 SciDex: The Natural Products Identifier, 1st ed; Wiley-VCH: Weinheim, Germany, 2007. [Google Scholar]
- Buckingham, J. Dictionary of Natural Products on CD-ROM, 16.2 ed; Chapman and Hall: London, UK, 2008. [Google Scholar]
- Laursen, JB; Nielsen, J. Phenazine natural products: Biosynthesis, synthetic analogues, and biological activity. Chem Rev 2004, 104, 1663–1686. [Google Scholar]
| Phylogenetic group | Number of strains | ||
|---|---|---|---|
| Analyzed | PCR amplification phzE gene positive | Producing phenazines in culture | |
| Actinobacteriaa | 76 | 11 | 11 |
| Bacteroidetes | 2 | 0 | 0 |
| Firmicutes | 28 | 1 | 0 |
| Alphaproteobacteria | 21 | 2 | 0 |
| Betaproteobacteria | 2 | 0 | 0 |
| Gammaproteobacteriab | 39 | 3 | 3 |
| In total | 168 | 17 | 14 |
| Strain no. | Next relative type strain and acc.-no.a | Phylumb | Sequence lengthc | Related phenazine gene, acc.-no.d, similarity and producer |
|---|---|---|---|---|
| Strains used as DSM 1042T | Positive control Streptomyces cinnamonensis DSM 1042T; DQ462657 | A | 127 | phzE; CAL34110; 100%; S. cinnamonenis |
| DSM 6698T | Pseudomonas chlororaphis subsp. aureofaciens DSM 6698T; AY509898 | GP | 139 | phzE: ADP21173; 100% P. chlororaphis phzF: ADP21174; 100% P. chlororaphis |
| DSM 19603T | Pseudomonas chlororaphis subsp. aurantiaca DSM 19603T; DQ682655 | GP | 137 | phzE: ADP21173; 98% P. chlororaphis phzF: ADP21174 49% P. chlororaphis |
| DSM 50083T | Pseudomonas chlororaphis subsp. chlororaphis DSM 50083T; Z76673 | GP | 125 | phzE; AAF17499; 92% P. chlororaphis phzF: AAF17500; 99% P. chlororaphis |
| Environmental | Isolates | |||
| AB108 | Pseudovibrio ascidiaceicola F423T; AB175663 | AP | 144 | phzE; CAL34110; 95%; S. cinnamonenis |
| HB117 | Streptomyces fulvorobeus LMG 19901T; AJ781331 | A | 141 | phzE; AAF17499; 73%; P. chlororaphis |
| HB122 | Streptomyces luridiscabiei S63T AF361784 | A | 141 | phzE; AAF17499; 74%; P. chlororaphis |
| HB202 | Streptomyces mediolani LMG 20093T; AJ781354 | A | 91 | phzE; NP_252903; 84%; P. aeruginosa |
| HB253 | Micromonospora matsumotoense IMSNU 22003T; AF152109 | A | 144 | phzB; AAF17496; 67%; P. chlororaphis |
| HB254 | Micromonospora matsumotoense IMSNU 22003T; AF152109 | A | 140 | phzE; AAF17499; 73%; P. chlororaphis |
| HB291 | Streptomyces fulvorobeus LMG 19901T; AJ781331 | A | 140 | phzE; AAF17499; 73%; P. chlororaphis |
| LB066 | Kiloniella laminariae LD81T; AM749667 | AL | 132 | phzE; CAL34110; 91%; S. cinnamonenis |
| LB114 | Streptomyces flavogriseus DSM 40323T; AJ494864 | A | 141 | phzE; AAF17499; 79%; P. chlororaphis |
| LB129 | Streptomyces fimicarius ISP 5322T; AY999784 | A | 145 | phzB; AAF17496; 75%; P. chlororaphis |
| LB150 | Streptomyces luridiscabiei S63T; AF361784 | A | 132 | phzB; AAF17496; 74%; P. chlororaphis |
| LB151 | Streptomyces griseus ATCC51928T; AF112160 | A | 133 | phzE; AAF17499; 65%; P. chlororaphis |
| Strain no. | Next relative type strain | [M+] | UV absorption maxima (nm)a | Dereplication of phenazines |
|---|---|---|---|---|
| Strains used as DSM 1042T | positive control Streptomyces cinnamonensis DSM 1042T | 206 | 327, 249, 212 | no hit in database |
| 224 | 371, 249, 215 | phenazine-1-carboxylic acid [41] | ||
| 268 | 375, 256, 223 | phenazine-1,6-dicarboxylic acid [15] | ||
| 292 | 371, 254, 214 | endophenazine A [15] c | ||
| 306 | 387, 269, 211 | no hit in database | ||
| 308 | 372, 249, 212 | endophenazine C [15] | ||
| 322 | 375, 256, 223 | endophenazine B [15]c | ||
| 336 | 372, 249, 212 | no hit in database | ||
| DSM 6698T | Pseudomonas chlororaphis subsp. aureofaciens DSM 6698T | 196 | 368, 257, 219 | 2-hydroxy-phenazine [42] |
| 224 | 371, 249, 215 | phenazine-1-carboxylic acid [41] | ||
| DSM 19603 T | Pseudomonas chlororaphis subsp. aurantiaca DSM 19603T | 196 | 368, 257, 219 | 2-hydroxy-phenazine [42]c |
| 224 | 371, 249, 215 | phenazine-1-carboxylic acid [41]c | ||
| DSM 50083 T | Pseudomonas chlororaphis subsp. chlororaphis DSM 50083T | 223 | 370, 248, 213 | chlororaphin [43] |
| Environmental | Isolates | |||
| HB117 | Streptomyces fulvorobeus LMG 19901T | 494 | 370(br), 274, 224 | Senacarcin A |
| 512 | 370(br), 275, 230 | saphenyl ester D [29] | ||
| HB122 | Streptomyces luridiscabiei S63T | 492 | 376, 275, 235sh | saphenyl ester D [29] |
| 496 | 438sh, 383(br), 276, 227 | no hit in database | ||
| 498 | 419sh, 393-325, 289, 253sh, 220 | no hit in database | ||
| 508 | 376, 275, 235sh | no hit in database | ||
| 510 | 430(br), 325, 224 | derivative of aestivophoenin C [44] | ||
| 512 | 432(br), 327, 226 | aestivophoenin C [44] | ||
| HB202 | Streptomyces mediolani LMG 20093T | 396 | 368, 364sh, 351sh, 252, 218 | streptophenazines E [39]c |
| 410 | 371, 364sh, 354sh, 252, 213 | streptophenazines C [39] | ||
| 410 | 368, 364sh, 351sh. 252, 218 | streptophenazines D [39]c | ||
| 424 | 367, 363sh, 350sh, 252, 215 | streptophenazines A [39] | ||
| 424 | 368, 364sh, 351sh, 252, 218 | streptophenazines B [39]c | ||
| 438 | 368, 364sh, 353sh, 252, 215 | streptophenazines F [39]c | ||
| 438 | 368, 363sh, 351sh, 252, 214 | streptophenazines G [39] | ||
| 440 | 368, 363sh, 352sh, 252, 215 | streptophenazines H [39] | ||
| HB253 | Micromonospora matsumotoense IMSNU 22003T | 260 | 458, 302sh, 261, 232 | no hit in database |
| 465 | 362sh, 345, 299, 221 | no hit in database | ||
| 566 | 362sh, 345, 299, 221 | no hit in database | ||
| HB254 | Micromonospora matsumotoense IMSNU 22003T | 451 | 361, 343, 352, 301, 223 | no hit in database |
| HB291 | Streptomyces fulvorobeus LMG 19901T | 492 | 376, 275, 235sh | saphenyl ester D [29] |
| 496 | 438sh, 383(br), 276, 227 | no hit in database | ||
| 498 | 419sh, 393-325, 289, 253sh, 220 | no hit in database | ||
| 508 | 376, 275, 235sh | no hit in database | ||
| 510 | 430(br), 325, 224 | derivative of aestivophoenin C [44] | ||
| 512 | 432(br), 327, 226 | aestivophoenin C [44] | ||
| LB114 | Streptomyces flavogriseus DSM 40323T | n.d.b | 370, 270, 244 | no hit in database |
| n.d.b | 419, 367, 305, 228 | no hit in database | ||
| LB129 | Streptomyces fimicarius ISP 5322T | 296 | 366, 249, 214 | phencomycin methyl ester [9] |
| 238 | 366, 249, 214 | 1-carboxymethyl phenazine | ||
| LB150 | Streptomyces luridiscabiei S63T | 510 | sh401, 378, 274, 227 | no hit in database |
| LB151 | Streptomyces griseus ATCC 51928T | 510 | sh401, 378, 274, 227 | no hit in database |
| Primer | Sequence | Function | Ref. |
|---|---|---|---|
| 27f | 5′-GAGTTTGATCCTGGCTCAG-3′ | PCR of the 16S rRNA gene | [54] |
| 1492r | 5′-GGTTACCTTGTTACGACTT-3′ | PCR of the 16S rRNA gene | [54] |
| 534r | 5′-ATTACCGCGGCTGCTGG-3′ | Sequencing of the 16S rRNA gene | [55] |
| 342f | 5′-TACGGGAGGCAGCAG-3′ | sequencing of the 16S rRNA gene | [55] |
| 790f | 5′-GATACCCTGGTAGTCC-3′ | sequencing of the 16S rRNA gene | [50] |
| phzEf | 5′-GAAGGCGCCAACTTCGTYATCAA-3′ | PCR and sequencing of phzE gene | this study |
| phzEr | 5′-GCCYTCGATGAAGTACTCGGTGTG-3′ | PCR and sequencing of phzE gene | this study |
| Ps_up1 | 5′-ATCTTCACCCCGGTCAACG-3′ | PCR and sequencing of phzF gene | [21] |
| Ps_low1 | 5′-CCRTAGGCCGGTGAGAAC-3′ | PCR and sequencing of phzF gene | [21] |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Schneemann, I.; Wiese, J.; Kunz, A.L.; Imhoff, J.F. Genetic Approach for the Fast Discovery of Phenazine Producing Bacteria. Mar. Drugs 2011, 9, 772-789. https://doi.org/10.3390/md9050772
Schneemann I, Wiese J, Kunz AL, Imhoff JF. Genetic Approach for the Fast Discovery of Phenazine Producing Bacteria. Marine Drugs. 2011; 9(5):772-789. https://doi.org/10.3390/md9050772
Chicago/Turabian StyleSchneemann, Imke, Jutta Wiese, Anna Lena Kunz, and Johannes F. Imhoff. 2011. "Genetic Approach for the Fast Discovery of Phenazine Producing Bacteria" Marine Drugs 9, no. 5: 772-789. https://doi.org/10.3390/md9050772




