Pyropia yezoensis Protein Supplementation Prevents Dexamethasone-Induced Muscle Atrophy in C57BL/6 Mice
Abstract
:1. Introduction
2. Results
2.1. PYCP Supplementation Attenuates DEX-Induced Loss of Body Weight
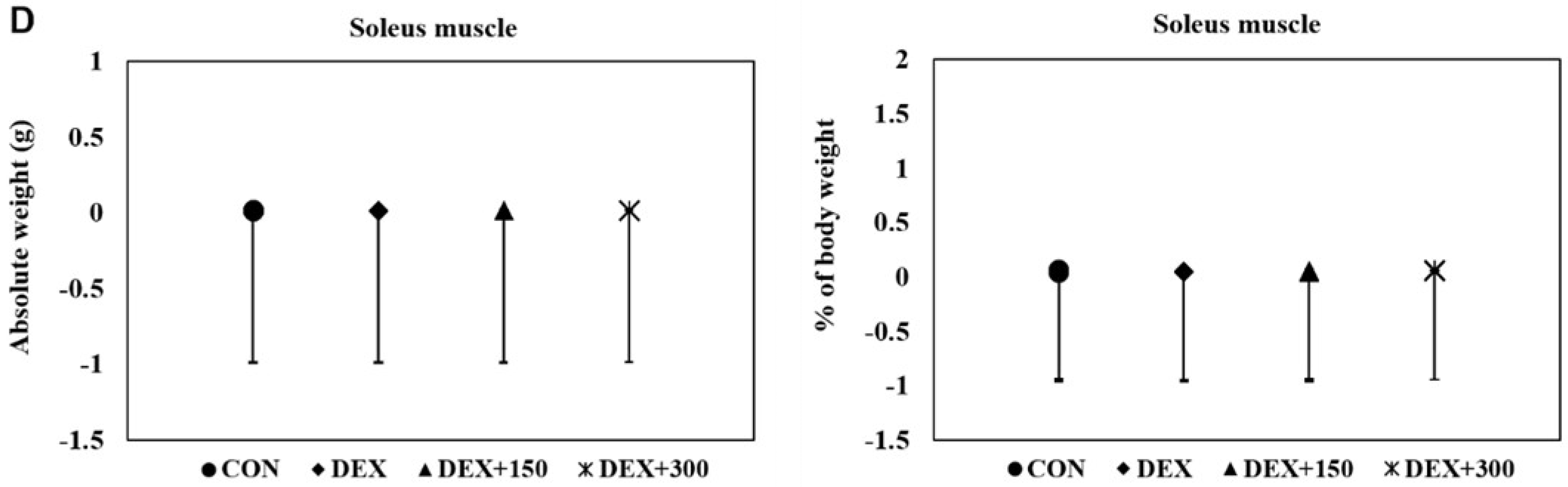
2.2. PYCP Supplementation Attenuates DEX-Induced Loss of Calf Thickness and Muscle Weight
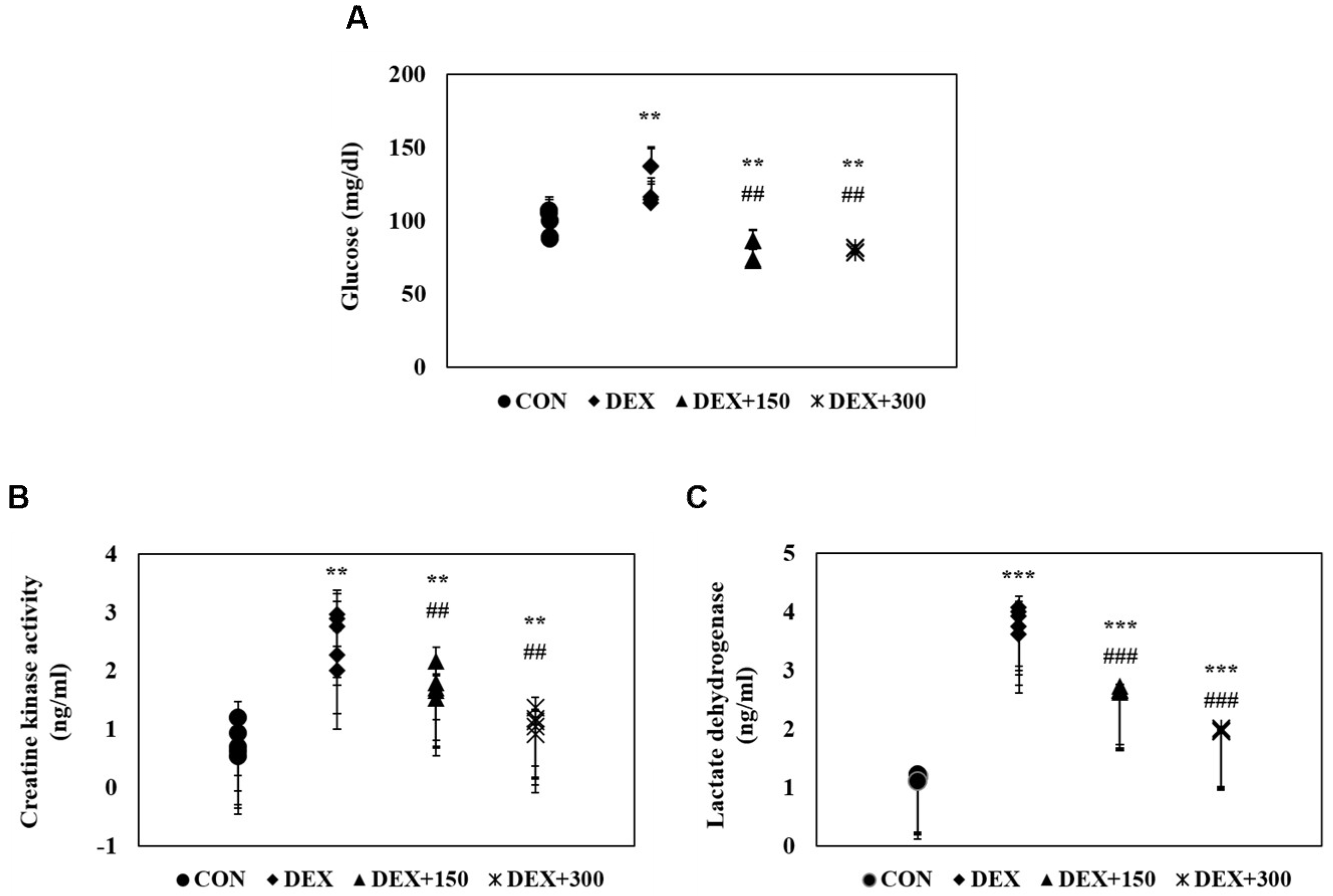
2.3. Changes in Serum Biochemistry in Mice with DEX-Induced Muscle Atrophy
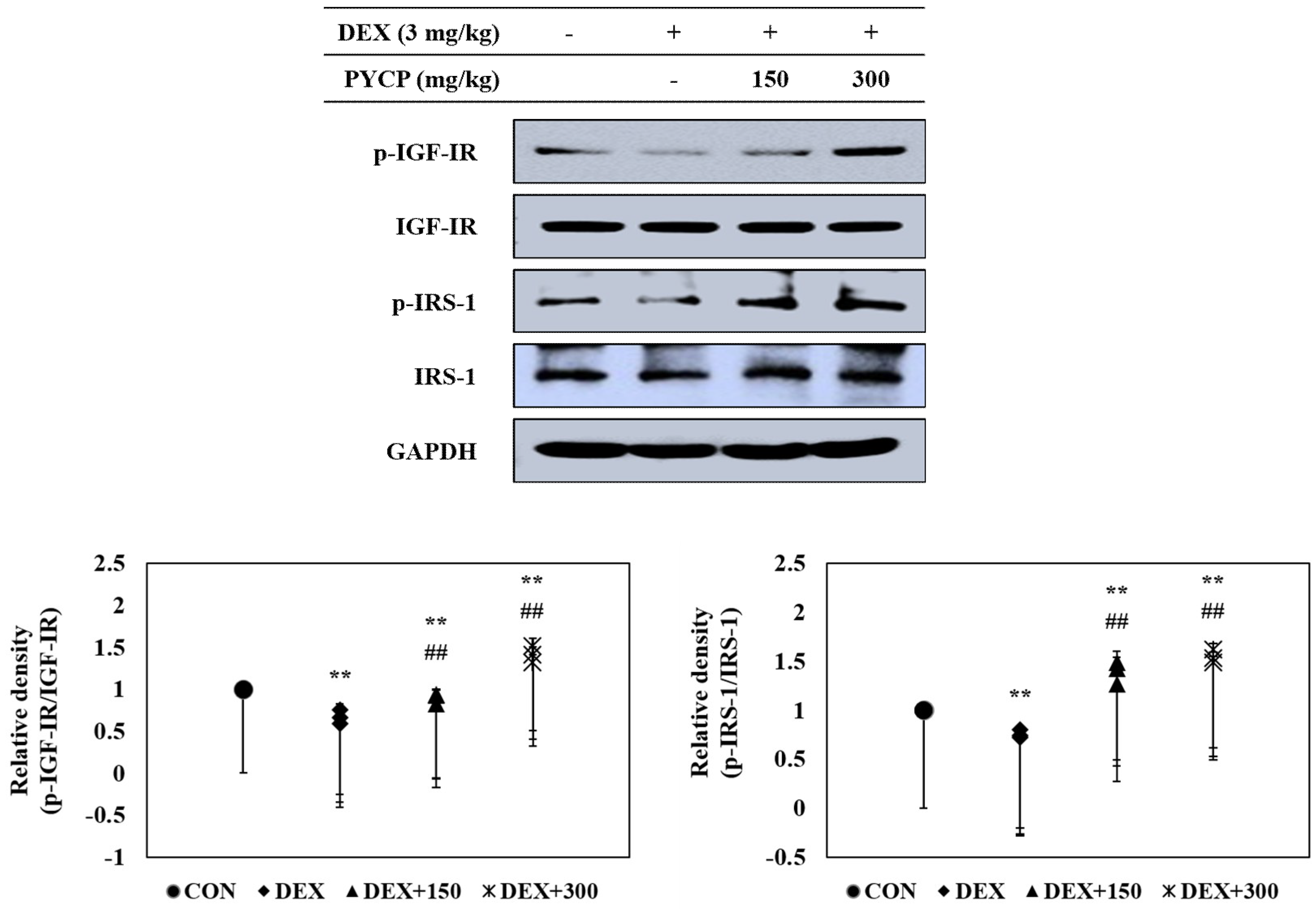
2.4. PYCP Supplementation Upregulates the Decrease in DEX-Induced IGF-IR and IRS-1 Phosphorylation in Gastrocnemius Muscle
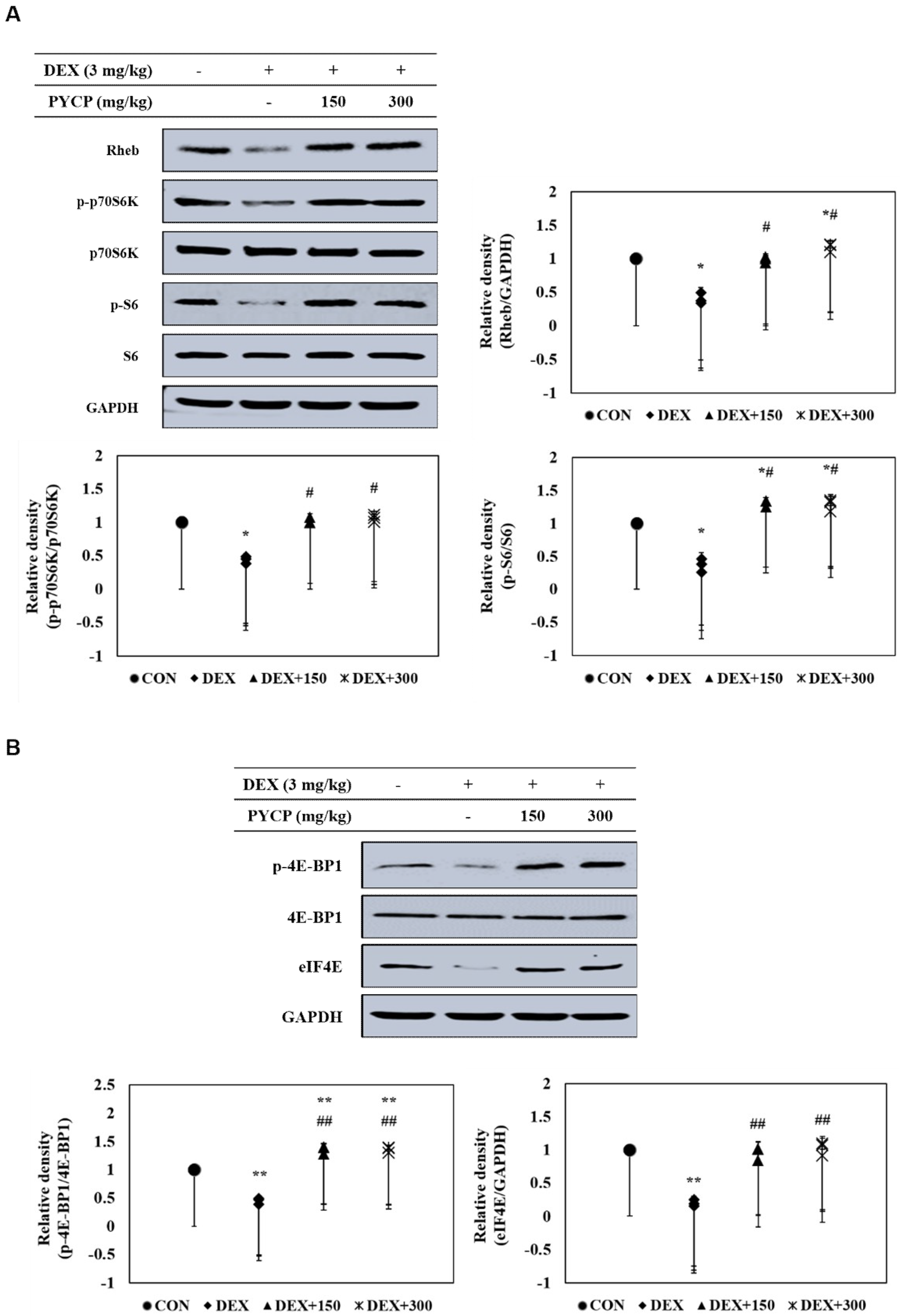
2.5. PYCP Supplementation Upregulates the Decrease in the DEX-Induced Akt/mTORC1 Signaling Pathway in Gastrocnemius Muscle
2.6. PYCP Supplementation Upregulates the Decrease in DEX-Induced p70S6K and 4E-BP1 Phosphorylation in Gastrocnemius Muscle
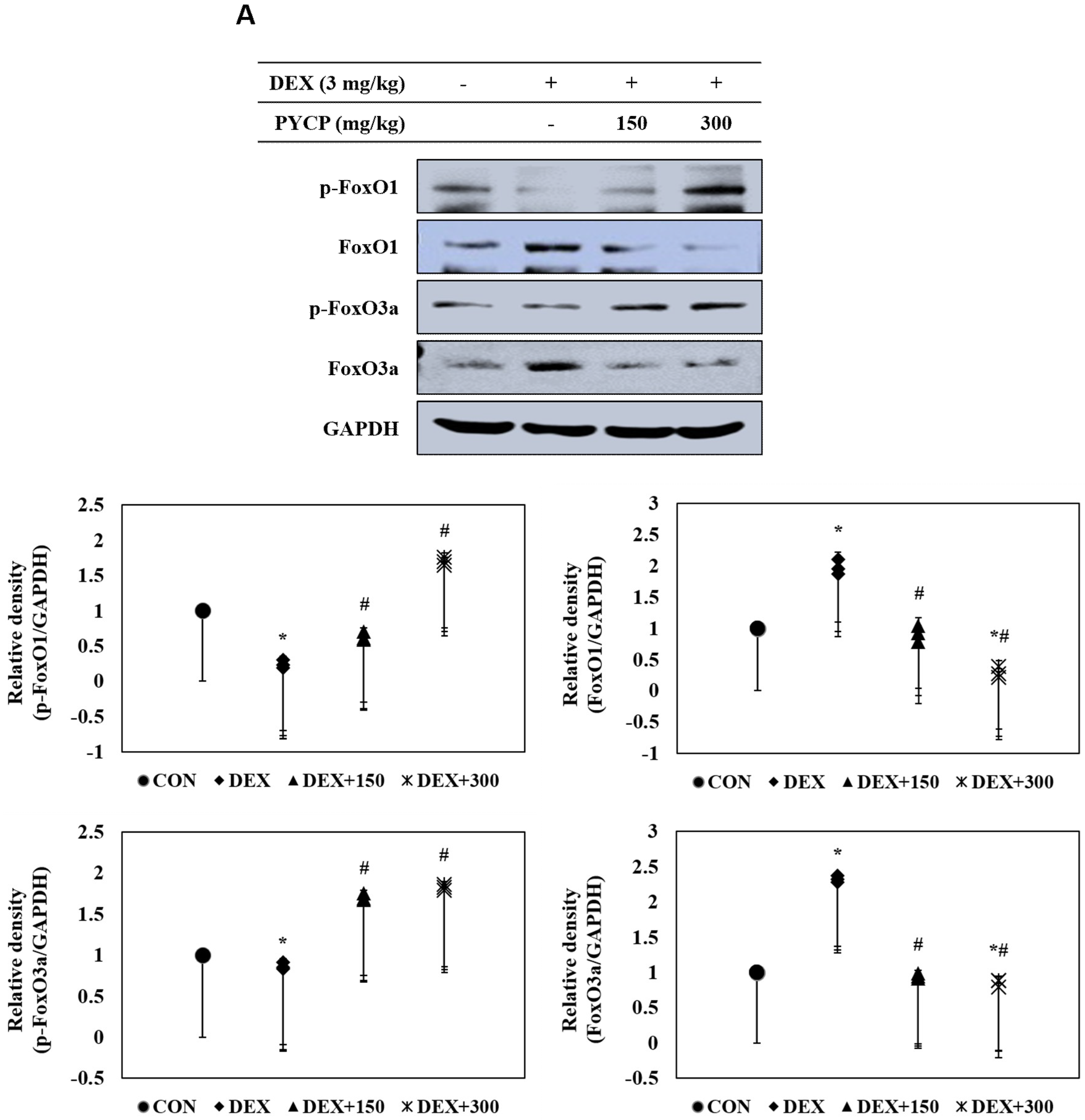
2.7. PYCP Supplementation Downregulates the Increase in DEX-Induced FoxO1 and FoxO3a Activation in Gastrocnemius Muscle
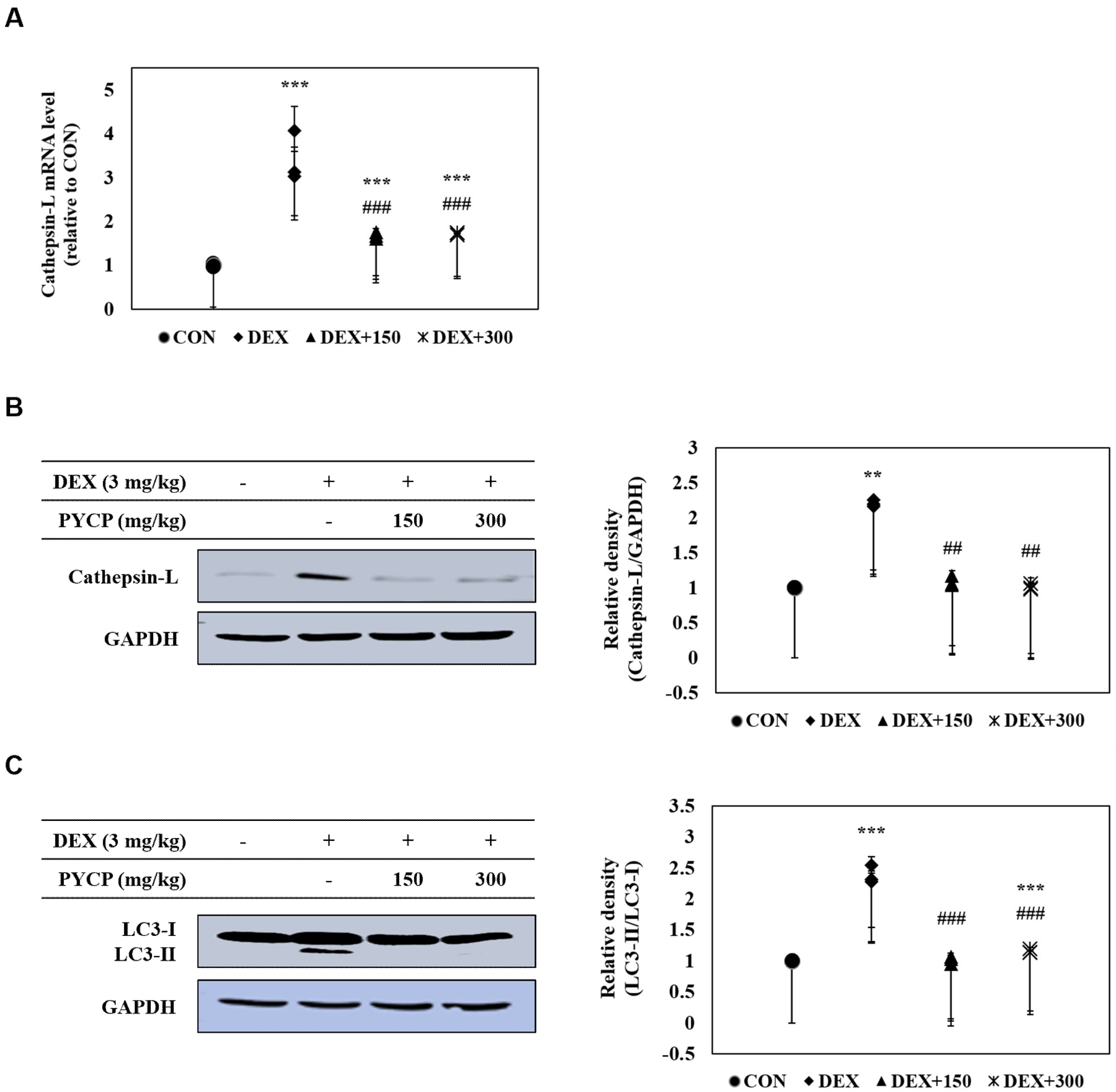
2.8. PYCP Supplementation Inhibits the DEX-Induced Activation of the Ubiquitin-Proteasome System in Gastrocnemius Muscle
2.9. PYCP Supplementation Inhibits the Activation of the DEX-Induced Autophagy-Lysosome System in Gastrocnemius Muscle
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. Experimental Scheme
4.3. Measurement of Body Weight and Calf Thickness
4.4. Measurement of Gastrocnemius, Soleus, and Tibialis Anterior Muscle Weight
4.5. Measurement of Glucose Uptake
4.6. Measurement of Creatine Kinase Activity
4.7. Measurement of Lactate Dehydrogenase Activity
4.8. Preparation of Total Cell Lysates
4.9. Preparation of Cytosolic and Nuclear Extracts
4.10. Western Blot Analysis
4.11. Real-Time Polymerase Chain Reaction
4.12. Measurement of 20S Proteasome Activity
4.13. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Na, E.; Dharmarajan, K.; et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, C.; Russo, T.L.; Sandoval, M.C.; Dentillo, A.A.; Couto, M.A.; Durigan, J.L.; Salvini, T.F. Joint inflammation alters gene and protein expression and leads to atrophy in the tibialis anterior muscle in rats. Am. J. Phys. Med. Rehabil. 2011, 90, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Scientific rationale for inhaled combination therapy with long-acting beta2-agonists and corticosteroids. Eur. Respir. J. 2002, 19, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J.; Adcock, I.M. Transcription factors and asthma. Eur. Respir. J. 1998, 12, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Cole, C.H. Postnatal glucocorticosteroid therapy for treatment and prevention of neonatal chronic lung disease. Expert Opin. Investig. Drugs 2000, 9, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Dardevet, D.; Sornet, C.; Taillandier, D.; Savary, I.; Attaix, D.; Grizard, J. Sensitivity and protein turnover response to glucocorticoids are different in skeletal muscle from adult and old rats. Lack of regulation of the ubiquitin-proteasome proteolytic pathway in aging. J. Clin. Investig. 1995, 96, 2113–2119. [Google Scholar] [CrossRef] [PubMed]
- Mitch, W.E.; Goldberg, A.L. Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. N. Engl. J. Med. 1996, 335, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Batchelor, T.T.; Taylor, L.P.; Thaler, H.T.; Posner, J.B.; Deangelis, L.M. Steroid myopathy in cancer patients. Neurology 1997, 48, 1234–1238. [Google Scholar] [CrossRef] [PubMed]
- Baehr, L.M.; Furlow, J.D.; Bodine, S.C. Muscle sparing in muscle RING finger 1 null mice: Response to synthetic glucocorticoids. J. Physiol. 2011, 19, 4759–4776. [Google Scholar] [CrossRef] [PubMed]
- Castillero, E.; Alamdari, N.; Lecker, S.; Hasselgren, P.O. Suppression of atrogin-1 and MuRF1 prevents dexamethasone-induced atrophy of cultured myotubes. Metabolism 2013, 62, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Glass, D.J. Skeletal muscle hypertrophy and atrophy signaling pathways. Int. J. Biochem. Cell Biol. 2003, 37, 1974–1984. [Google Scholar] [CrossRef] [PubMed]
- Sartorelli, V.; Fulco, M. Molecular and cellular determinants of skeletal muscle atrophy and hypertrophy. Sci. STKE 2004, 2004, re11. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Mammucari, C. Regulation of skeletal muscle growth by the IGF-I-Akt/PKB pathway: Insights from genetic models. Skelet. Muscle 2011, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Mavalli, M.D.; Digirolamo, D.J.; Fan, Y.; Riddle, R.C.; Campbell, K.S.; Van, G.T.; Frank, S.J.; Sperling, M.A.; Esser, K.A.; Bamman, M.M.; et al. Distinct growth hormone receptor signaling modes regulate skeletal muscle development and insulin sensitivity in mice. J. Clin. Investig. 2010, 120, 4007–4020. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; Lee, M.N.; Yang, Y.R.; Jeong, H.; Ghim, J.; Noh, J.; Kim, J.Y.; Ryu, D.R.; Park, S.H.; Song, P.K.; et al. C1-Ten is a protein tyrosine phosphatase of IRS-1, regulating IRS-1 stability and muscle atrophy. Mol. Cell Biol. 2013, 33, 1608–1620. [Google Scholar] [CrossRef] [PubMed]
- Nakao, R.; Hirasaka, K.; Goto, J.; Ishidoh, K.; Yamada, C.; Ohno, A.; Okumura, Y.; Nonaka, I.; Yasutomo, K.; Baldwin, K.M.; et al. Ubiquitin ligase Cn1-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Mol. Cell Biol. 2009, 29, 4798–4811. [Google Scholar] [CrossRef] [PubMed]
- Kukreti, H.; Amuthavalli, K.; Harikumar, A.; Sathiyamoorthy, S.; Feng, P.Z.; Anantharaj, R.; Tan, S.L.K.; Lokireddy, S.; Bonala, S.; Sriram, S.; et al. Muscle-specific microRNA1 (miR1) targets heat shock protein 70 (HSP70) during dexamethasone-mediated atrophy. J. Biol. Chem. 2013, 288, 6663–6678. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kubica, N.; Ellisen, L.W.; Jefferson, L.S.; Kimball, S.R. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. J. Biol. Chem. 2006, 281, 39128–39134. [Google Scholar] [CrossRef] [PubMed]
- Mammucari, C.; Schiaffino, S.; Sandri, M. Downstream of Akt: FoxO3 and mTOR in the regulation of autophagy in skeletal muscle. Autophagy 2008, 4, 524–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Southgate, R.J.; Neill, B.; Prelovsek, O.; El-Osta, A.; Kamei, Y.; Miura, S.; Exaki, O.; McLoughlin, T.J.; Zhang, W.; Unterman, T.G.; et al. FoxO1 regulates the expression of 4E-BP1 and inhibits mTOR signaling in mammalian skeletal muscle. J. Biol. Chem. 2007, 282, 21176–21186. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M. Signaling in muscle atrophy and hypertrophy. Physiology 2008, 23, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Stitt, T.N.; Drujan, D.; Clarke, B.A.; Panaro, F.; Timofeyva, Y.; Kline, W.O.; Gonzalez, M.; Yancopoulos, G.D.; Glass, D.J. The IGF-I/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 2004, 14, 395–403. [Google Scholar] [CrossRef]
- Cho, J.E.; Fournier, M.; Da, X.; Lewis, M.I. Time course expression of Foxo transcription factors in skeletal muscle following corticosteroid administration. J. Appl. Physiol. 2010, 108, 137–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilson, H.; Schakman, O.; Combaret, L.; Lause, P.; Grobet, L.; Attaiz, D.; Ketelslegers, J.M.; Thissen, T.P. Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology 2007, 148, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Imae, M.; Fu, Z.; Yoshida, A.; Noguchi, T.; Kato, H. Nutritional and hormonal factors control the gene expression of FoxOs, the mammalian homologues of DAF-16. J. Mol. Endocrinol. 2003, 30, 253–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagoe, R.T.; Lecker, S.H.; Gomes, M.; Goldberg, A.L. Patterns of gene expression in atrophying skeletal muscles: Response to food deprivation. FASEB J. 2002, 16, 1697–1712. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M.; Sandri, C.; Gilbert, A.; Skurk, C.; Calabria, E.; Picard, A.; Walsh, K.; Schiaffino, S.; Lecker, S.H.; Goldberg, A.L. FoxO transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004, 117, 399–412. [Google Scholar] [CrossRef]
- Hasselgren, P.O. Glucocorticoids and muscle catabolism. Curr. Opin. Clin. Nutr. Metab. 1999, 2, 201–205. [Google Scholar] [CrossRef]
- Niwa, K. Genetic analysis of artificial green and red mutants of Porphyra yezoensis Ueda (Bangiales, Rhodophyta). Aquaculture 2012, 308, 6–12. [Google Scholar] [CrossRef]
- Qu, W.; Ma, H.; Pan, Z.; Luo, L.; Wang, Z.; He, R. Preparation and antihypertensive activity of peptides from Porphyra yezoensis. Food Chem. 2010, 123, 14–20. [Google Scholar] [CrossRef]
- Kim, E.Y.; Choi, Y.H.; Choi, C.G.; Nam, T.J. Effects of the cyclophilin-type peptidylprolyl cis-trans isomerase from Pyropia yezoensis against hydrogen peroxide-induced oxidative stress in HepG2 cells. Mol. Med. Rep. 2017, 15, 4132–4138. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Kim, Y.M.; Park, S.J.; Kim, I.H.; Nam, T.J. Protective effect of Porphyra yezoensis glycoprotein on D-galactosamine-induced cytotoxicity in Hepa 1c1c7 cells. Mol. Med. Rep. 2015, 11, 3914–3919. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Kim, I.H.; Kim, Y.M.; Nam, T.J. Pyropia yezoensis glycoprotein regulates antioxidant status and prevents hepatotoxicity in rat model of d-galactosamin/lipopolysaccharide-induced acute liver failure. Mol. Med. Rep. 2016, 13, 3110–3114. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.; Gao, S.; Puppa, M.J.; Sato, S.; Welle, S.L.; Carson, J.A. Testosterone regulation of Akt/mTORC1/FoxO3a signaling in skeletal muscle. Mol. Cell. Endocrinol. 2013, 365, 174–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Du, R.; Yang, Y.Q.; Zhang, H.Q.; Li, Q.; Liu, L.; Guan, H.; Hou, J.; An, X.R. Dexamethasone-induced skeletal muscle atrophy was associated with upregulation of myostatin promoter activity. Res. Vet. Sci. 2013, 94, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, N.; Yoshikawa, N.; Ito, N.; Maruyama, T.; Suzuki, Y.; Takeda, S.I.; Nakae, J.; Tagata, Y.; Nishitani, S.; Takehana, K.; et al. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell. Metab. 2011, 13, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, J.J.; Wang, Z.Q.; Wang, N.; Wu, Z.Y. Value of muscle enzyme measurement in evaluating different neuro-muscular diseases. Clin. Chim. Acta 2012, 413, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Park, H.; Cho, S.; Lee, M. Vitamin D3 supplementation modulates inflammatory responses from the muscle damage induced by high-intensity exercise in SD rats. Cytokine 2013, 63, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Zaho, J.; Brault, J.J.; Schild, A.; Goldberg, A.L. Coordinate activation of autophagy and the proteasome pathway by FoxO transcription factor. Autophagy 2008, 4, 378–380. [Google Scholar] [CrossRef] [Green Version]
- Clarke, B.A.; Drujan, D.; Willis, M.S.; Murphy, L.O.; Corpina, R.A.; Burova, E.; Rakhilin, S.V.; Stitt, T.N.; Patterson, C.; Latres, E.; et al. The E3 ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007, 6, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Li, B.G.; Hasselgren, P.O.; Fang, C.H. Insulin-like growth factor-I inhibits dexamethasone-induced proteolysis in culture L6 myotubes through PI3K/Akt/GSK-3β and PI3K/Akt/mTOR-dependent mechanisms. Int. J. Biochem. Cell Biol. 2005, 37, 2207–2216. [Google Scholar] [CrossRef] [PubMed]
- Troncoso, R.; Paredes, F.; Parra, V.; Gatica, D.; Vasquez-Trincado, C.; Quiroga, C.; Bravo-Sagua, R.; Lopez-Crisosto, C.; Rodriguez, A.E.; Oyarzun, A.P.; et al. Dexamethasone-induced autophagy mediates muscle atrophy through mitochondrial clearance. Cell Cycle 2014, 13, 2281–2295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.K.; Kim, I.H.; Choi, Y.H.; Nam, T.J. A peptide from Porphyra yezoensis stimulates the proliferation of IEC-6 cells by activating the insulin-like growth factor I receptor signaling pathway. Int. J. Mol. Med. 2015, 35, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Ryu, J.A.; Kim, I.H.; Choi, Y.H.; Nam, T.J. Induction of apoptosis by a peptide from Porphyra yezoensis: Regulation of the insulin-like growth factor-I receptor signaling pathway in MCF-7 cells. Int. J. Oncol. 2014, 45, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Frost, R.A.; Lang, C.H. Regulation of insulin-like growth factor-I in skeletal muscle and muscle cells. Minerva Endocrinol. 2003, 28, 53–73. [Google Scholar] [PubMed]
- Inder, W.J.; Jang, C.; Obeyesekere, V.R.; Alford, F.P. Dexamethasone administration inhibits skeletal muscle expression of the androgen receptor and IGF-implications for steroid-induced myopathy. Clin. Endocrinol. 2010, 73, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Rommel, C.; Bodine, S.C.; Clarke, B.A.; Rossman, R.; Nunez, L.; Stitt, T.N.; Yancopoulos, G.D.; Glass, D.J. Mediation of IGF-I induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR/GSK3 pathway. Nat. Cell Biol. 2001, 3, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Jellyman, J.K.; Martin-Gronery, M.S.; Cripps, R.L.; Giussani, D.A.; Ozamme, S.E.; Shen, Q.W.; Du, M.; Fowden, A.L.; Forhead, A.J. Effects of cortisol and dexamethasone on insulin signaling pathway in skeletal muscle of the ovine fetus during late gestation. PLoS ONE 2012, 7, e52363. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Shimizu, N.; Yoshikawa, N. Role of skeletal muscle glucocorticoid receptor in systemic energy homeostasis. Exp. Cell Res. 2017, 360, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Britto, F.A.; Begue, G.; Rossano, B.; Docquier, A.; Vernus, B.; Sar, C.; Ferry, A.; Bonnieu, A.; Ollendorff, V.; Favier, F.B. REDD1 deletion prevents dexamethasone-induced skeletal muscle atrophy. Am. J. Physiol. Endocrinol. Metab. 2014, 307, E983–E993. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Kimball, S.R.; Jahn, L.A.; Barrett, E.J. Glucocorticoids modulate amino acid-induced translation initiation in human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2004, 287, E275–E281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, D.L.; Unterman, T.G. Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am. J. Physiol. 2007, 292, C188–C199. [Google Scholar] [CrossRef] [PubMed]
- Kamei, Y.; Miura, S.; Suzuki, M.; Kai, Y.; Mizukami, J.; Taniguchi, T.; Mochida, K.; Hata, T.; Matsuda, J.; Aburatani, H.; et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 2004, 279, 41114–41123. [Google Scholar] [CrossRef] [PubMed]
- Zaho, W.; Pan, J.; Zhao, Z.; Wu, Y.; Bauman, W.A.; Cardozo, C.P. Testosterone protects against dexamethasone-induced muscle atrophy, protein degradation and MAFbx upregulation. J. Steroid Biochem. Mol. Biol. 2008, 110, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Giron, M.D.; Vilchez, J.D.; Shreeram, S.; Salto, R.; Manzano, M.; Cabrera, E.; Campos, N.; Edens, N.K.; Rueda, R.; Lopez-Pedrosa, J.M. β-Hydroxy- β-methylbutyrate (HMB) normalizes dexamethasone-induced autophagy-lysosomal pathway in skeletal muscle. PLoS ONE 2015, 10, e0117520. [Google Scholar] [CrossRef] [PubMed]
- Zaho, W.; Qin, W.; Pan, J.; Wu, Y.; Bauman, W.A.; Cardozo, C. Dependence of dexamethasone-induced Akt/FOXO1 signaling, upregulation of MAFbx, and protein catabolism upon the glucocorticoid receptor. Biochem. Biophys. Res. Commun. 2009, 378, 668–672. [Google Scholar] [CrossRef] [PubMed]





| Group | ||||
|---|---|---|---|---|
| CON | DEX | DEX + PYCP150 | DEX + PYCP300 | |
| Basal BW (g) | 24.13 ± 0.69 | 24.04 ± 0.41 | 24.69 ± 0.97 | 24.17 ± 0.84 |
| Final BW (g) | 25.77 ± 0.41 | 24.35 ± 0.57 * | 25.22 ± 1.01 # | 25.47 ± 0.65 # |
| Antibody | Manufacturer and Catalog No. | Species of Origin | Dilution Rate |
|---|---|---|---|
| 4E-BP1 | Santa Cruz Biotechnology: sc-9977 | Mouse | 1:1000 |
| Akt | Santa Cruz Biotechnology: sc-8312 | Rabbit | 1:1000 |
| ATG5 | Santa Cruz Biotechnology: sc-33210 | Rabbit | 1:1000 |
| ATG7 | Santa Cruz Biotechnology: sc-376212 | Mouse | 1:1000 |
| ATG12 | Santa Cruz Biotechnology: sc-271688 | Mouse | 1:1000 |
| Atrogin-1/MAFbx | Santa Cruz Biotechnology: sc-27645 | Goat | 1:2000 |
| β-actin | Santa Cruz Biotechnology: sc-47778 | Mouse | 1:1000 |
| BECN1 | Santa Cruz Biotechnology: sc-11427 | Rabbit | 1:1000 |
| Cathepsin-L | Santa Cruz Biotechnology: sc-6498 | Goat | 1:1000 |
| eIF4E | Santa Cruz Biotechnology: sc-514875 | Mouse | 1:1000 |
| FoxO1 | Santa Cruz Biotechnology: sc-374427 | Mouse | 1:500 |
| FoxO3a | Santa Cruz Biotechnology: sc-9813 | Goat | 1:1000 |
| GAPDH | Santa Cruz Biotechnology: sc-25778 | Rabbit | 1:1000 |
| IGF-IR | Santa Cruz Biotechnology: sc-713 | Rabbit | 1:1000 |
| IRS-1 | Santa Cruz Biotechnology: sc-560 | Rabbit | 1:1000 |
| Lamin B | Santa Cruz Biotechnology: sc-377000 | Mouse | 1:1000 |
| LC3-I/II | Cell Signaling: #4108S | Rabbit | 1:1000 |
| mTOR | Santa Cruz Biotechnology: sc-8319 | Rabbit | 1:1000 |
| MuRF1 | Santa Cruz Biotechnology: sc-27642 | Goat | 1:2000 |
| p62/SQSTM1 | Cell Signaling: #5114S | Rabbit | 1:1000 |
| p-4E-BP1 | Santa Cruz Biotechnology: sc-293124 | Mouse | 1:1000 |
| p70S6K | Santa Cruz Biotechnology: sc-8418 | Mouse | 1:1000 |
| p-Akt | Santa Cruz Biotechnology: sc-135650 | Mouse | 1:500 |
| p-FoxO1 | Cell Signaling: #9461S | Rabbit | 1:500 |
| p-FoxO3a | Cell Signaling: #9466S | Rabbit | 1:1000 |
| p-IGF-IR | Santa Cruz Biotechnology: sc-101703 | Rabbit | 1:1000 |
| p-IRS-1 | Santa Cruz Biotechnology: sc-17200 | Goat | 1:1000 |
| p-mTOR | Santa Cruz Biotechnology: sc-293132 | Mouse | 1:1000 |
| p-p70S6K | Santa Cruz Biotechnology: sc-8416 | Mouse | 1:1000 |
| p-S6 | Santa Cruz Biotechnology: sc-293144 | Mouse | 1:1000 |
| Raptor | Santa Cruz Biotechnology: sc-81537 | Mouse | 1:1000 |
| Rheb | Santa Cruz Biotechnology: sc-271509 | Mouse | 1:1000 |
| Rictor | Santa Cruz Biotechnology: sc-81538 | Mouse | 1:1000 |
| S6 | Santa Cruz Biotechnology: sc-74459 | Mouse | 1:1000 |
| Gene | Accession No. | Sequence (5′–3′) | Amplicon (bp) |
|---|---|---|---|
| Atrogin-1/MAFbx | NM_026346.3 | F: ATGCACACTGGTGCAGAGAG R: TGTAAGCACACAGGCAGGTC | 168 |
| Cathepsin-L | M20495.1 | F: GACCGGGACAACCACTGTG R: CCCATCAATTCACGACAGGAT | 61 |
| MuRF1 | DQ229108.1 | F: TGTCTGGAGGTCGTTTCCG R: GTGCCGGTCCATGATCACTT | 59 |
| GAPDH | NM_008084.3 | F: ACTCCACTCACGGCAAATTCA R: CGCTCCTGGAAGATGGTGAT | 91 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.-K.; Choi, J.-W.; Choi, Y.H.; Nam, T.-J. Pyropia yezoensis Protein Supplementation Prevents Dexamethasone-Induced Muscle Atrophy in C57BL/6 Mice. Mar. Drugs 2018, 16, 328. https://doi.org/10.3390/md16090328
Lee M-K, Choi J-W, Choi YH, Nam T-J. Pyropia yezoensis Protein Supplementation Prevents Dexamethasone-Induced Muscle Atrophy in C57BL/6 Mice. Marine Drugs. 2018; 16(9):328. https://doi.org/10.3390/md16090328
Chicago/Turabian StyleLee, Min-Kyeong, Jeong-Wook Choi, Youn Hee Choi, and Taek-Jeong Nam. 2018. "Pyropia yezoensis Protein Supplementation Prevents Dexamethasone-Induced Muscle Atrophy in C57BL/6 Mice" Marine Drugs 16, no. 9: 328. https://doi.org/10.3390/md16090328





