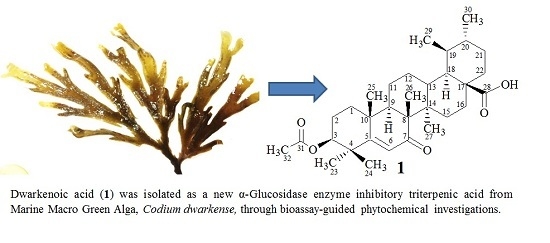
New α-Glucosidase Inhibitory Triterpenic Acid from Marine Macro Green Alga Codium dwarkense Boergs
Abstract
:1. Introduction
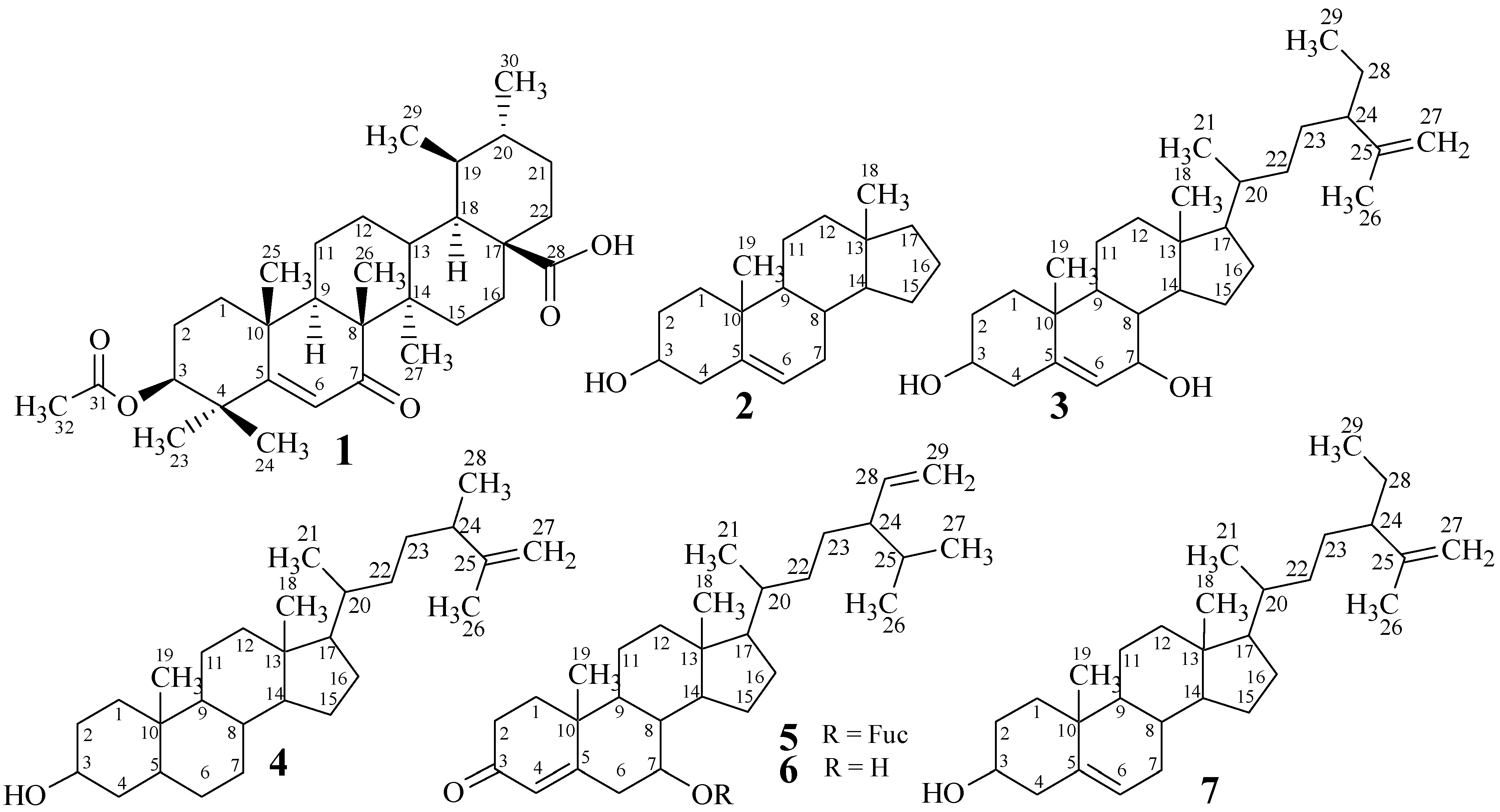
2. Results and Discussion
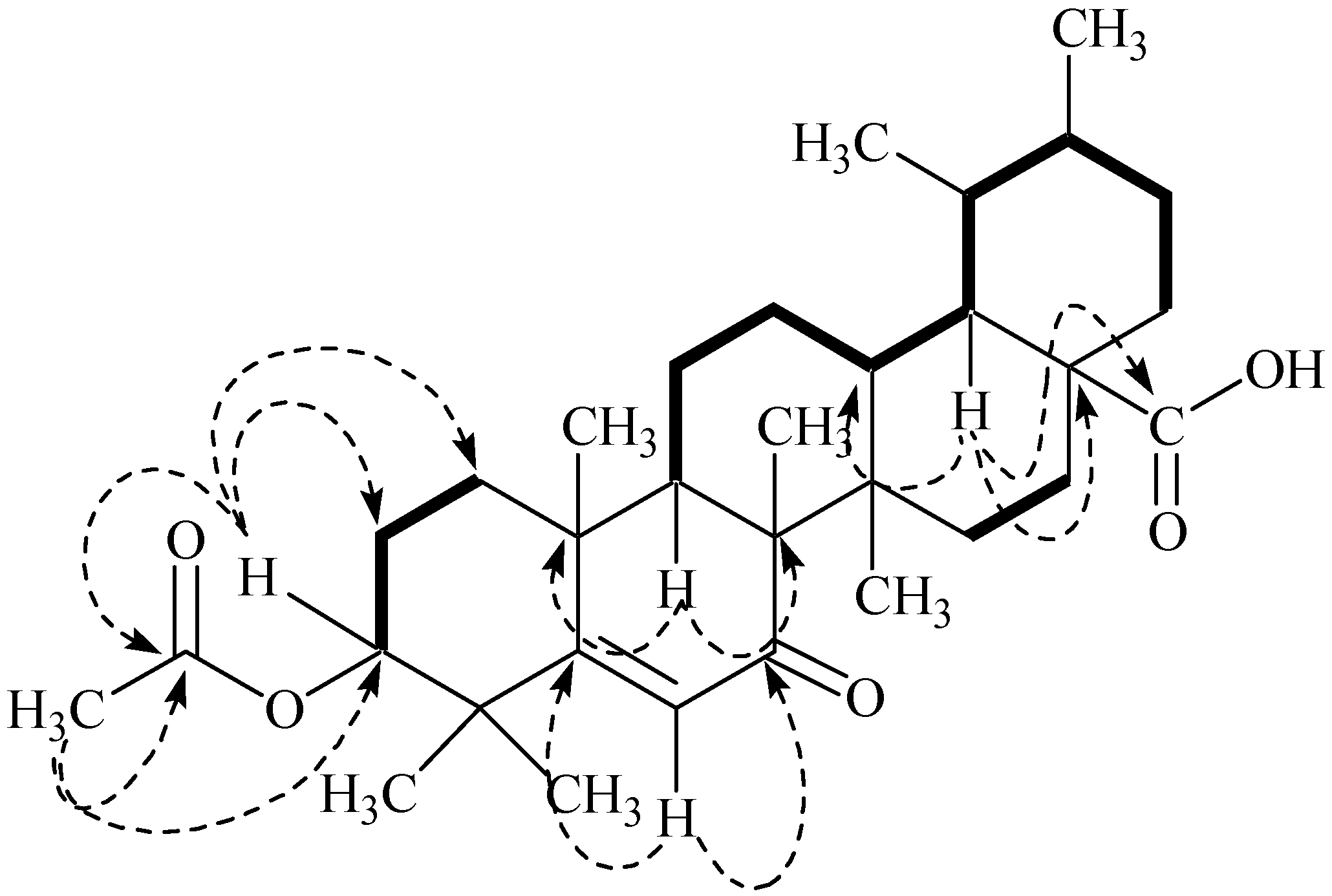
2.1. Structure Elucidation
| C. No. | 13C (δ) | 1H (δ) | Multiplicity | Key HMBC |
|---|---|---|---|---|
| 1 | 40.9 | 1.50, m | CH2 | |
| 2 | 23.5 | 2.11, m; 2.21, m | CH2 | |
| 3 | 73.1 | 5.29, br s | CH | 1,2,32 |
| 4 | 33.9 | - | C | |
| 5 | 164.9 | - | C | |
| 6 | 130.5 | 5.54, s | CH | 5,7 |
| 7 | 199.4 | - | C | |
| 8 | 45.1 | - | C | |
| 9 | 60.1 | 2.39, m | CH | 8,10 |
| 10 | 37.3 | - | C | |
| 11 | 27.2 | 1.87, m; 1.98, m | CH2 | |
| 12 | 29.7 | 1.89, m; 2.08, m | CH2 | |
| 13 | 50.4 | 1.38, m | CH | |
| 14 | 46.3 | - | C | |
| 15 | 32.8 | 1.45, m; 1.66, m | CH2 | |
| 16 | 34.6 | 2.29, m; 2.51, m | CH2 | |
| 17 | 43.8 | - | C | |
| 18 | 59.0 | 1.54, m | CH | 13,17,28 |
| 19 | 39.2 | 1.37, m | CH | |
| 20 | 39.3 | 0.92, m | CH | |
| 21 | 30.9 | 1.27, m; 1.42, m | CH2 | |
| 22 | 27.5 | 0.99, m, 1.20, m | CH2 | |
| 23 | 28.9 | 0.81, s | CH3 | 4 |
| 24 | 21.1 | 0.93, s | CH3 | 4 |
| 25 | 20.5 | 1.33, s | CH3 | |
| 26 | 13.3 | 1.14, s | CH3 | |
| 27 | 18.3 | 1.18, s | CH3 | |
| 28 | 179.1 | - | C | |
| 29 | 17.4 | 0.78, d, J = 6.5 Hz | CH3 | |
| 30 | 23.8 | 1.23, d, J = 6.5 Hz | CH3 | |
| 31 | 170.2 | - | COCH3 | |
| 32 | 21.3 | 2.07, s | COCH3 | 3,31 |

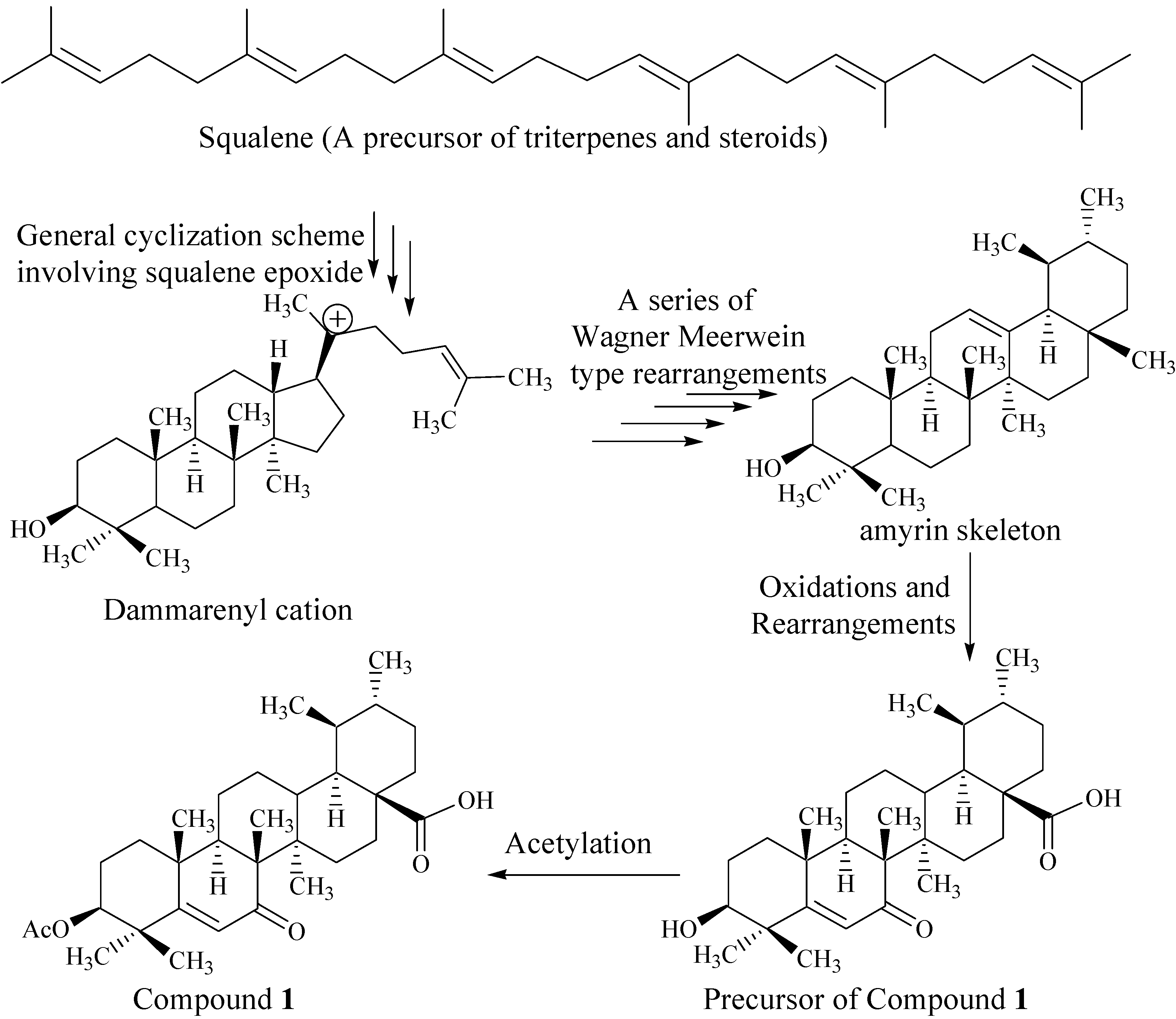
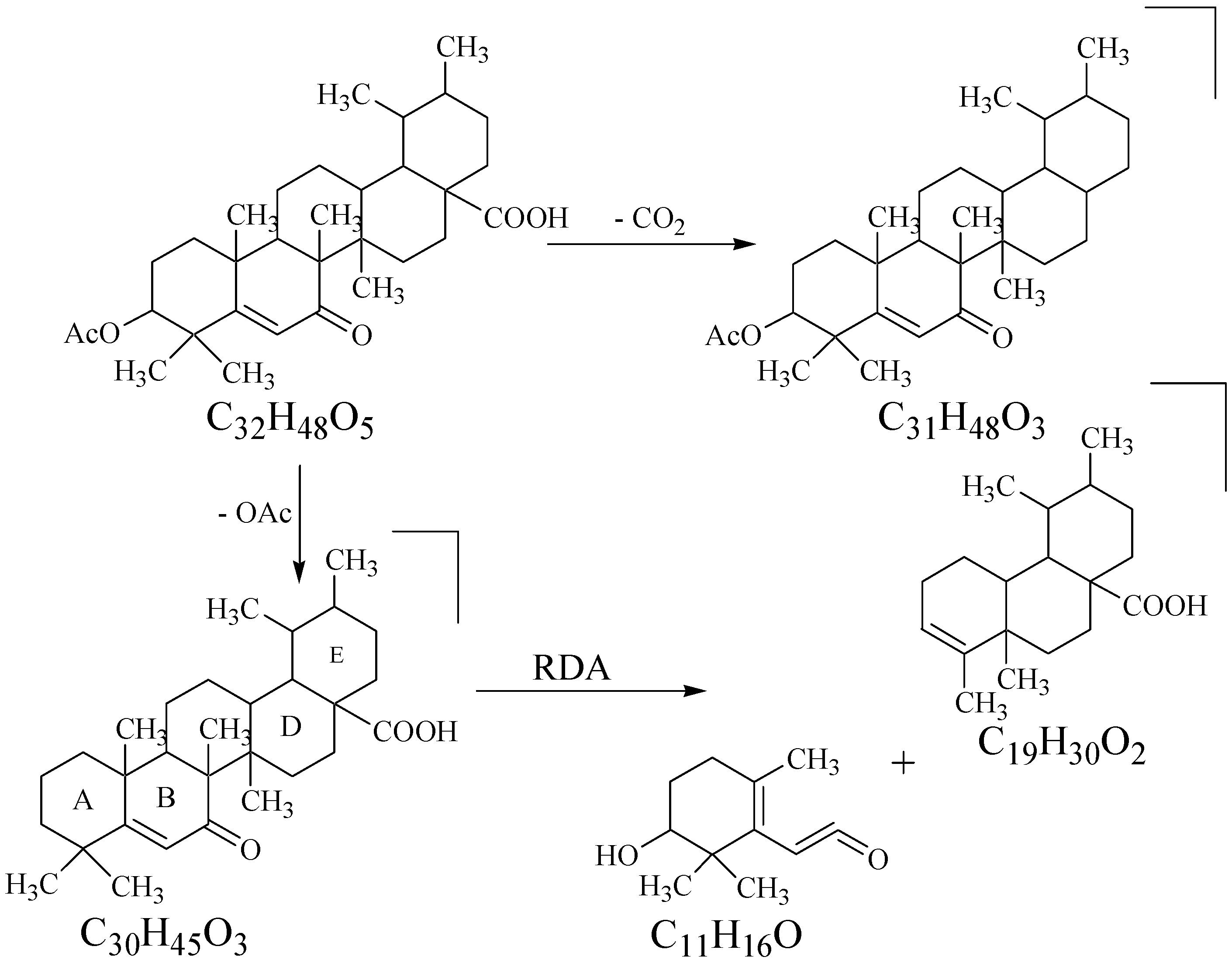
Biogenesis of Compound 1
2.2. Biological Evaluations
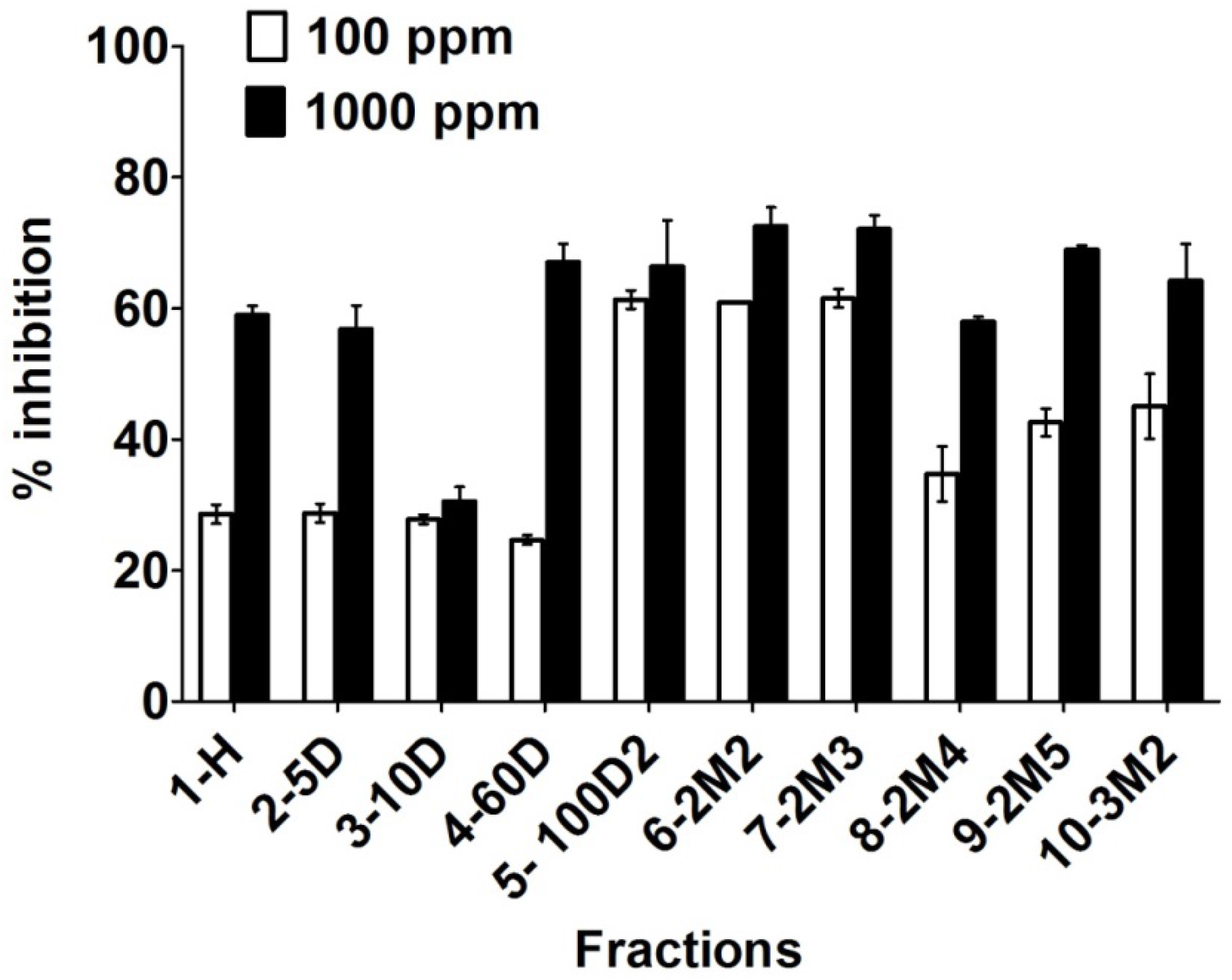
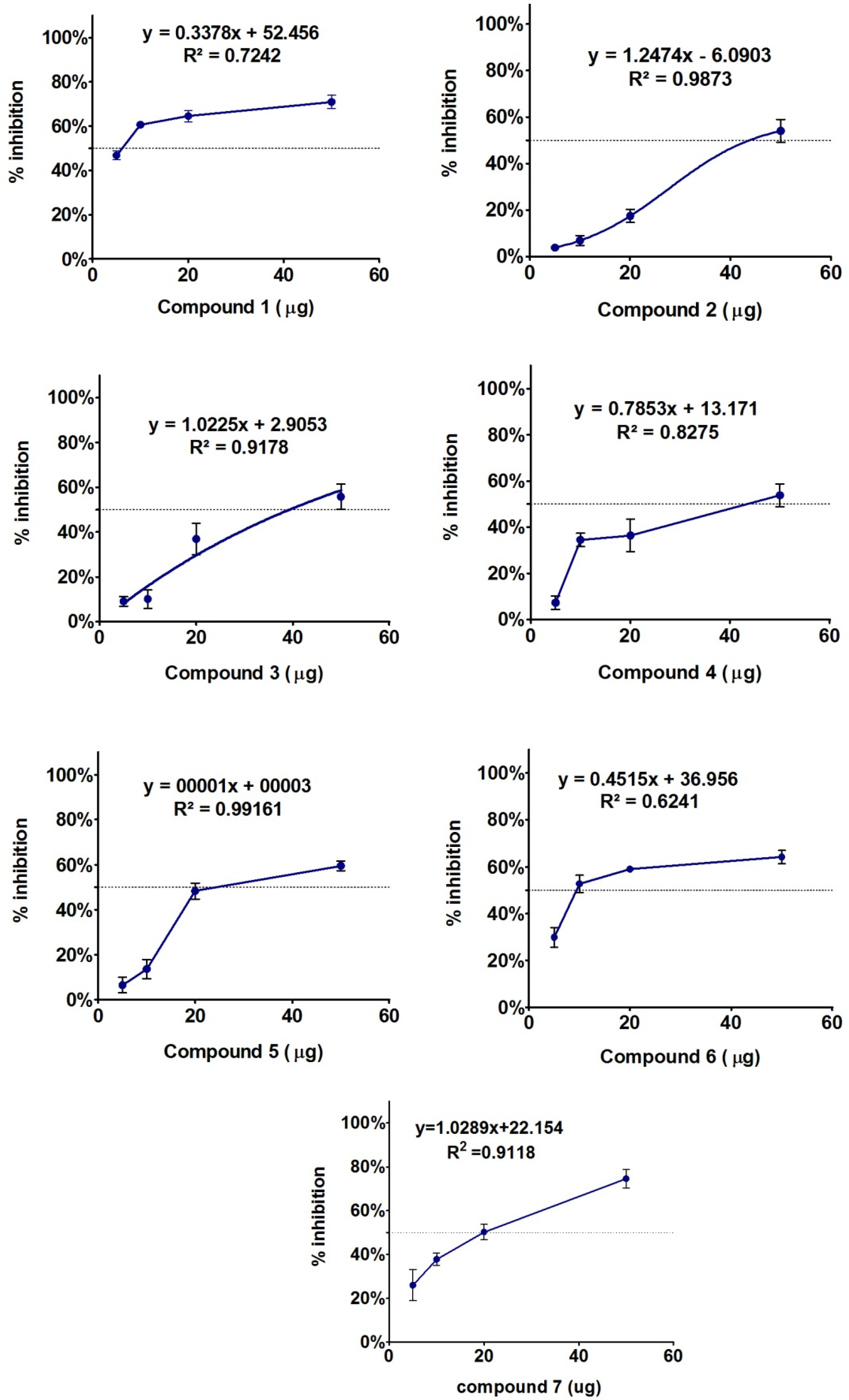
3. Conclusions
4. Experimental Section
4.1. General
4.2. Plant Material
4.3. Extraction and Isolation
Dwarkenoic Acid (1)
4.4. Enzyme Inhibition Assay
Supplementary Information
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kim, S.K.; Karagozlu, M.Z. Marine algae: Natural product source for gastrointestinal cancer treatment. Adv. Food Nutr. Res. 2011, 64, 225–233. [Google Scholar] [PubMed]
- Kim, S.K.; Ta, Q.V. Potential beneficial effects of marine algal sterols on human health. Adv. Food Nutr. Res. 2011, 64, 191–198. [Google Scholar] [PubMed]
- Kim, A.D.; Lee, Y.; Kang, S.H.; Kim, G.Y.; Kim, H.S.; Hyun, J.W. Cytotoxic effect of clerosterol isolated from Codium fragile on A2058 Human Melanoma Cells. Mar. Drugs 2013, 11, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Sheu, J.H.; Liaw, C.C.; Duh, C.Y. Oxygenated clerosterols isolated from the marine alga Codium arabicum. J. Nat. Prod. 1995, 58, 1521–1526. [Google Scholar] [CrossRef]
- Rubinstein, I.; Goad, L.J. Sterols of the siphonous marine alga Codium fragile. Phytochemistry 1974, 13, 481–484. [Google Scholar] [CrossRef]
- Ahmed, V.U.; Alyia, R.; Perveen, S.; Shameel, M. A sterol glycoside from marine green alga Codium iyengarii. Phytochemistry 1992, 31, 1429–1431. [Google Scholar] [CrossRef]
- Ahmed, V.U.; Alyia, R.; Perveen, S.; Shameel, M. Sterols from marine green alga Codium decorticatum. Phytochemistry 1993, 33, 1189–1192. [Google Scholar] [CrossRef]
- Schroeder, G.; Rohmer, M.; Beck, J.P.; Anton, R. 7-Oxo-, 7α-hydroxy- and 7β-hydroxysterols from Euphorbia fischeriana. Phytochemistry 1980, 19, 2213–2215. [Google Scholar] [CrossRef]
- Heltzel, C.E.; Gunatilaka, A.A.; Kingston, D.G.; Hofmann, G.A.; Johnson, R.K. Synthesis and structure-activity relationships of cytotoxic 7-hydroxy sterols. J. Nat. Prod. 1994, 57, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.J.; Jurd, K.M.; Blunden, G.; Paoletti, S.; Zanetti, F. Anticoagulant activity of a proteoglycan in extracts of Codium fragile ssp. atlanticum. J. Appl. Phycol. 1990, 2, 357–361. [Google Scholar] [CrossRef]
- Aliya, R.; Shameel, M.; Perveen, S.; Ali, M.S.; Usmanghani, K.; Ahmad, V.U. Acyclic diterpene alcohols from four algae of bryopsidophyceae and their toxicity. Pak. J. Mar. Sci. 1994, 3, 15–24. [Google Scholar]
- Ali, M.S.; Jahangir, M.; Saleem, M.; Pervez, M.K.; Hameed, S.; Ahmad, V.U. Metabolites of marine algae collected from Karachi-coasts of Arabian Sea. Nat. Prod. Sci. 2000, 6, 61–65. [Google Scholar]
- Kumar, S.; Narwal, S.; Kumar, V.; Prakash, O. α-Glucosidase inhibitors from plants: A natural approach to treat diabetes. Pharmacogn. Rev. 2011, 5, 19–29. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Health Estimates: Deaths by Cause, Age, Sex and Country, 2000–2012; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- Kim, K.Y.; Nam, K.A.; Kurihara, H.; Kim, S.M. Potent α-glucosidase inhibitors purified from the red alga Grateloupia elliptica. Phytochemistry 2008, 69, 2820–2825. [Google Scholar] [CrossRef] [PubMed]
- Kalra, S. Alpha glucosidase inhibitors. J. Pak. Med. Assoc. 2014, 64, 474–476. [Google Scholar] [PubMed]
- Wehmeier, U.F.; Piepersberg, W. Biotechnology and molecular biology of the alpha-glucosidase inhibitor acarbose. Appl. Microbiol. Biotechnol. 2004, 63, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, H. Pharmacology of α-glucosidase inhibition. Eur. J. Clin. Investig. 1994, 24, 3–10. [Google Scholar]
- Li, X.; Fan, X.; Han, L.; Lou, Q.; Yan, X.; Zhang, Y. Screening for alpha-Glucosidase Inhibitors from the Macroalgal Extracts. Chin. J. Mar. Drugs 2002, 21, 8–11. [Google Scholar]
- Siddhanta, A.K.; Shanmugam, M.; Mody, K.H.; Goswami, A.M.; Ramavat, B.K. Sulphated polysaccharides of Codium dwarkense Boergs. from the west coast of India: Chemical composition and blood anticoagulant activity. Int. J. Biol. Macromol. 1999, 26, 151–154. [Google Scholar] [CrossRef]
- Shanmugam, M.; Mody, K.H.; Siddhanta, A.K. Blood anticoagulant sulphated polysaccharides of the marine green algae Codium dwarkense (Boergs.) and C. tomentosum (Huds.) Stackh. Ind. J. Exp. Biol. 2001, 39, 365–370. [Google Scholar]
- Shanmugam, M.; Mody, K.H.; Oza, R.M.; Ramavat, B.K. Blood anticoagulant activity of a green marine alga Codium dwarkense (Codiaceae, Chlorophyta) in relation to its growth stages. Ind. J. Mar. Sci. 2001, 30, 49–52. [Google Scholar]
- Sun, G.; Zhang, X.; Xu, X.; Yang, J.; Zhong, M.; Yuan, J. A new triterpene from the plant of Uncaria macrophylla. Molecules 2012, 17, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D. The biosynthesis of steroids and triterpenoids. Nat. Prod. Rep. 1998, 15, 653–696. [Google Scholar] [CrossRef]
- Dewick, P.M. Medicinal Natural Products. A Biosynthetic Approach; John Wiley & Sons: Chichester, UK, 2001; pp. 212–236. [Google Scholar]
- Mbaze, L.M.; Poumale, H.M.; Wansi, J.D.; Lado, J.A.; Khan, S.N.; Iqbal, M.C.; Nqadjui, B.T.; Laatsch, H. alpha-glucosidase inhibitory pentacyclic triterpenes from the stem bark of Fagara tessmannii (Rutaceae). Phytochemistry 2007, 68, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Razdan, T.K.; Kachroo, P.K.; Qurishi, M.A.; Kalla, A.K.; Waight, E.S. Unusual homologous long-chain alkanoic acid esters of lupeol from Koelpinia linearis. Phytochemistry 1996, 41, 1437–1438. [Google Scholar] [CrossRef]
- Oki, T.; Matsumoto, T.; Osajima, Y. Inhibitory effect of alpha-glucosidase inhibitors varies according to its origin. J. Agric. Food Chem. 1999, 47, 550–553. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, L.; Khan, A.L.; Al-Kharusi, L.; Hussain, J.; Al-Harrasi, A. New α-Glucosidase Inhibitory Triterpenic Acid from Marine Macro Green Alga Codium dwarkense Boergs. Mar. Drugs 2015, 13, 4344-4356. https://doi.org/10.3390/md13074344
Ali L, Khan AL, Al-Kharusi L, Hussain J, Al-Harrasi A. New α-Glucosidase Inhibitory Triterpenic Acid from Marine Macro Green Alga Codium dwarkense Boergs. Marine Drugs. 2015; 13(7):4344-4356. https://doi.org/10.3390/md13074344
Chicago/Turabian StyleAli, Liaqat, Abdul Latif Khan, Lubna Al-Kharusi, Javid Hussain, and Ahmed Al-Harrasi. 2015. "New α-Glucosidase Inhibitory Triterpenic Acid from Marine Macro Green Alga Codium dwarkense Boergs" Marine Drugs 13, no. 7: 4344-4356. https://doi.org/10.3390/md13074344