SD118-Xanthocillin X (1), a Novel Marine Agent Extracted from Penicillium commune, Induces Autophagy through the Inhibition of the MEK/ERK Pathway
Abstract
:1. Introduction
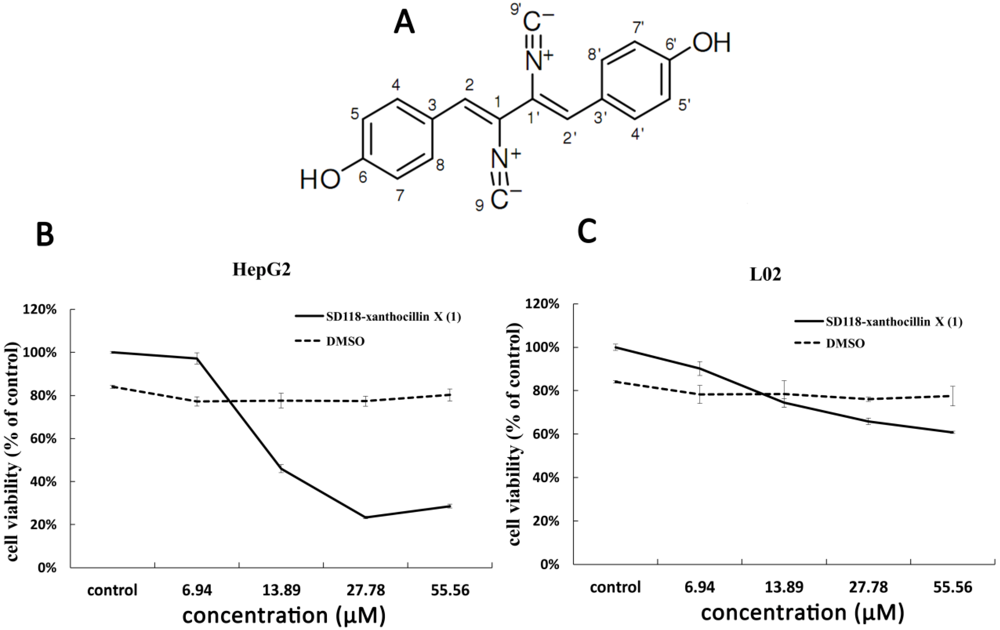
2. Results and Discussion
2.1. Cytotoxic Activity of SD118-Xanthocillin X (1) in HepG2 Cells
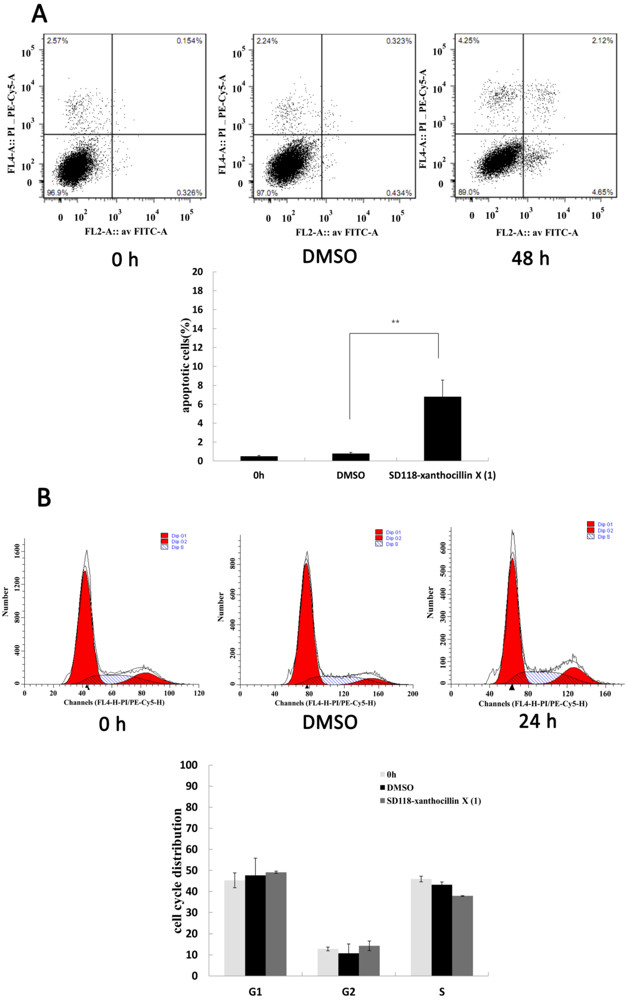
2.2. SD118-Xanthocillin X (1) Induces Apoptosis and Autophagy in HepG2 Cells

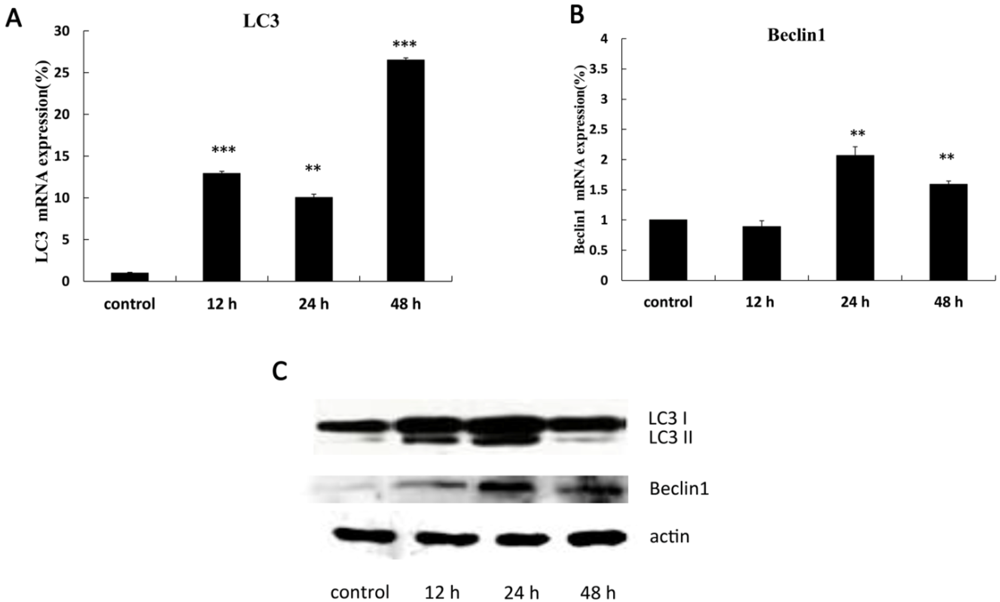
2.3. The Expression of Autophagy-Related Genes
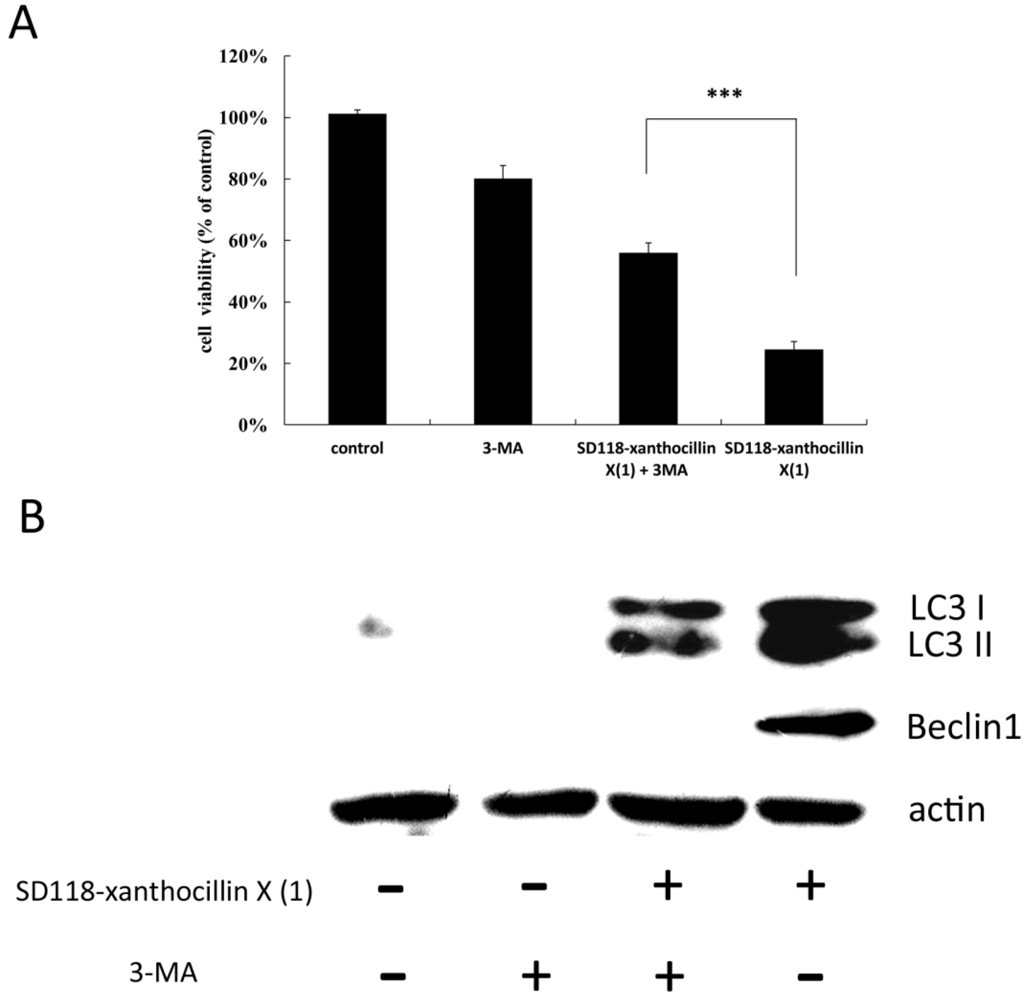
2.4. 3-MA Partially Attenuates the Autophagic Effect of SD118-Xanthocillin X (1)
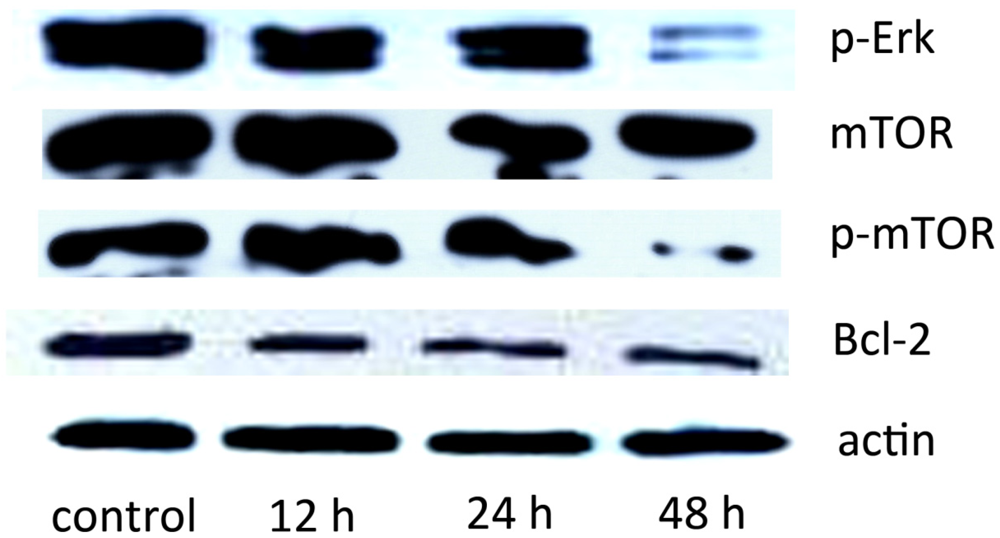
2.5. The Effect of SD118-Xanthocillin X (1) on Autophagy-Related Signaling Pathways
3. Experimental Section
3.1. Test Compound
3.2. Drug Preparation
3.3. Cell Lines and Growth Inhibition Assay
3.4. Flow Cytometry Analysis of Apoptosis
3.5. Transmission Electron Microscopy (TEM)
3.6. Reverse Transcription and Quantitative Real-Time PCR
3.7. Western Blotting
3.8. Statistical Analysis
4. Conclusion
Acknowledgments
References
- Naidu, S.S.; Naidu, S. Studies in Marine Quinone Chemistry; Rhodes Universityi: Grahamstown, South Africa, 2010. [Google Scholar]
- Nastrucci, C.; Russo, P. Anticancer drug discovery from the marine environment. Recent Pat. Anticancer Drug Discov. 2012, 7, 218–232. [Google Scholar] [CrossRef]
- Jordan, M.A.; Kamath, K.; Manna, T.; Okouneva, T.; Miller, H.P.; Davis, C.; Littlefield, B.A.; Wilson, L. The primary antimitotic mechanism of action of the synthetic halichondrin E7389 is suppression of microtubule growth. Mol. Cancer Ther. 2005, 4, 1086–1095. [Google Scholar] [CrossRef]
- Okouneva, T.; Azarenko, O.; Wilson, L.; Littlefield, B.A.; Jordan, M.A. Inhibition of centromere dynamics by eribulin (E7389) during mitotic metaphase. Mol. Cancer Ther. 2008, 7, 2003–2011. [Google Scholar] [CrossRef]
- Kuznetsov, G.; Towle, M.J.; Cheng, H.; Kawamura, T.; TenDyke, K.; Liu, D.; Kishi, Y.; Yu, M.J.; Littlefield, B.A. Induction of morphological and biochemical apoptosis following prolonged mitotic blockage by halichondrin B macrocyclic ketone analog E7389. Cancer Res. 2004, 64, 5760–5766. [Google Scholar]
- Towle, M.J.; Salvato, K.A.; Wels, B.F.; Aalfs, K.K.; Zheng, W.; Seletsky, B.M.; Zhu, X.; Lewis, B.M.; Kishi, Y.; Yu, M.J. Eribulin induces irreversible mitotic blockade: Implications of cell-based pharmacodynamics for in vivo efficacy under intermittent dosing conditions. Cancer Res. 2011, 71, 496–505. [Google Scholar]
- Klionsky, D.J. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007, 8, 931–937. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Gestwicki, J.E.; Murphy, L.O.; Klionsky, D.J. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007, 6, 304–312. [Google Scholar]
- Viola, G.; Bortolozzi, R.; Hamel, E.; Moro, S.; Brun, P.; Castagliuolo, I.; Ferlin, M.G.; Basso, G. MG-2477, a new tubulin inhibitor, induces autophagy through inhibition of the Akt/mTOR pathway and delayed apoptosis in A549 cells. Biochem. Pharmacol. 2012, 83, 16–26. [Google Scholar]
- Kanzawa, T.; Germano, I.; Komata, T.; Ito, H.; Kondo, Y.; Kondo, S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004, 11, 448–457. [Google Scholar] [CrossRef]
- Laane, E.; Tamm, K.P.; Buentke, E.; Ito, K.; Khahariza, P.; Oscarsson, J.; Corcoran, M.; Björklund, A.; Hultenby, K.; Lundin, J. Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death Differ. 2009, 16, 1018–1029. [Google Scholar] [CrossRef]
- Mizushima, N. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2491–2502. [Google Scholar] [CrossRef]
- Degenhardt, K.; Mathew, R.; Beaudoin, B.; Bray, K.; Anderson, D.; Chen, G. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006, 10, 51–64. [Google Scholar] [CrossRef]
- Mehrpour, M.; Esclatine, A.; Beau, I.; Codogno, P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010, 20, 748–762. [Google Scholar] [CrossRef]
- Shang, Z.; Li, X.; Meng, L.; Li, C.; Gao, S.; Huang, C.; Wang, B. Chemical profile of the secondary metabolites produced by a deep-sea sediment-derived fungus Penicillium commune SD-118. Chin. J. Oceanol. Limnol. 2012, 30, 305–314. [Google Scholar]
- Cao, Q.; Yu, C.; Xue, R.; Hsueh, W.; Pan, P.; Chen, Z.; Wang, S.; McNutt, M.; Gu, J. Autophagy induced by suberoylanilide hydroxamic acid in Hela S3 cells involves inhibition of protein kinase B and up-regulation of Beclin 1. Int. J. Biochem. Cell Biol. 2008, 40, 272–283. [Google Scholar]
- Wang, J.; Whiteman, M.W.; Lian, H.; Wang, G.; Singh, A.; Huang, D.; Denmark, T. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J. Biol. Chem. 2009, 284, 21412–21414. [Google Scholar]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2503–2518. [Google Scholar] [CrossRef]
- Baehrecke, E.H. Autophagy: Dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005, 6, 505–510. [Google Scholar] [CrossRef]
- Levine, B.; Deretic, V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007, 7, 767–777. [Google Scholar] [CrossRef]
- Munafó, D.B.; Colombo, M.I. A novel assay to study autophagy: Regulation of autophagosome vacuole size by amino acid deprivation. J. Cell Sci. 2001, 114, 3619–3629. [Google Scholar]
- Noda, T.; Ohsumi, Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998, 273, 3963–3966. [Google Scholar] [CrossRef]
- Wu, Y.T.; Tan, H.L.; Shui, G.; Bauvy, C.; Huang, Q.; Wenk, M.R.; Ong, C.N.; Codogno, P.; Shen, H.M. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 2010, 285, 10850–18061. [Google Scholar]
- Takeuchi, H.; Kondo, Y.; Fujiwara, K.; Kanzawa, T.; Aoki, H.; Mills, G.B.; Kondo, S. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 2005, 65, 3336–3346. [Google Scholar]
- Pattingre, S.; Levine, B. Bcl-2 inhibition of autophagy: A new route to cancer? Cancer Res. 2006, 66, 2885–2888. [Google Scholar] [CrossRef]
- Vara, J.Á.F.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef]
- Petiot, A.; Ogier-Denis, E.; Blommaart, E.F.C.; Meijer, A.J.; Codogno, P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J.Biol. Chem. 2000, 275, 992–998. [Google Scholar]
- Vieira, O.V.; Botelho, R.J.; Rameh, L.; Brachmann, S.M.; Matsuo, T.; Davidson, H.W.; Schreiber, A.; Backer, J.M.; Cantley, L.C.; Grinstein, S. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 2001, 155, 19–26. [Google Scholar] [CrossRef]
- Liang, X.H.; Yu, J.; Brown, K.; Levine, B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001, 61, 3443–3449. [Google Scholar]
- Shinojima, N.; Yokoyama, T.; Kondo, Y.; Kondo, S. Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy. Autophagy 2007, 3, 635–637. [Google Scholar]
- Aoki, H.; Takada, Y.; Kondo, S.; Sawaya, R.; Aggarwal, B.B.; Kondo, Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: Role of Akt and extracellular signal-regulated kinase signaling pathways. Mol. Pharmacol. 2007, 72, 29–39. [Google Scholar] [CrossRef]
- Zhu, J.; Horbinski, C.; Guo, F.; Watkins, S.; Uchiyama, Y.; Chu, C.T. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am. J. Pathol. 2007, 170, 75–86. [Google Scholar] [CrossRef]
- Corcelle, E.; Nebout, M.; Bekri, S.; Gauthier, N.; Hofman, P.; Poujeol, P.; Fénichel, P.; Mograbi, B. Disruption of autophagy at the maturation step by the carcinogen lindane is associated with the sustained mitogen-activated protein kinase/extracellular signal-regulated kinase activity. Cancer Res. 2006, 66, 6861–6870. [Google Scholar]
- Pattingre, S.; Bauvy, C.; Codogno, P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J. Biol. Chem. 2003, 278, 16667–16674. [Google Scholar]
- Subramaniam, S.; Unsicker, K. ERK and cell death: ERK1/2 in neuronal death. FEBS J. 2010, 277, 22–29. [Google Scholar] [CrossRef]
- Ferrell, J.E.; Bhatt, R.R. Mechanistic studies of the dual phosphorylation of mitogen-activated protein kinase. J. Biol. Chem. 1997, 272, 19008–19016. [Google Scholar] [CrossRef]
- Maiuri, M.C.; Zalckvar, E.; Kimchi, A.; Kroemer, G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007, 8, 741–752. [Google Scholar] [CrossRef]
- Zhou, F.; Yang, Y.; Xing, D. Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis. FEBS J. 2011, 278, 403–413. [Google Scholar] [CrossRef]
- Deeb, D.; Gao, X.; Dulchavsky, S.A.; Gautam, S.C. CDDO-Me induces apoptosis and inhibits Akt, mTOR and NF-κB signaling proteins in prostate cancer cells. Anticancer Res. 2007, 27, 3035–3044. [Google Scholar]
- Samples Availability: Available from the authors.
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhao, Y.; Chen, H.; Shang, Z.; Jiao, B.; Yuan, B.; Sun, W.; Wang, B.; Miao, M.; Huang, C. SD118-Xanthocillin X (1), a Novel Marine Agent Extracted from Penicillium commune, Induces Autophagy through the Inhibition of the MEK/ERK Pathway. Mar. Drugs 2012, 10, 1345-1359. https://doi.org/10.3390/md10061345
Zhao Y, Chen H, Shang Z, Jiao B, Yuan B, Sun W, Wang B, Miao M, Huang C. SD118-Xanthocillin X (1), a Novel Marine Agent Extracted from Penicillium commune, Induces Autophagy through the Inhibition of the MEK/ERK Pathway. Marine Drugs. 2012; 10(6):1345-1359. https://doi.org/10.3390/md10061345
Chicago/Turabian StyleZhao, Ying, Huan Chen, Zhuo Shang, Binghua Jiao, Bin Yuan, Weizhang Sun, Bingui Wang, Mingyong Miao, and Caiguo Huang. 2012. "SD118-Xanthocillin X (1), a Novel Marine Agent Extracted from Penicillium commune, Induces Autophagy through the Inhibition of the MEK/ERK Pathway" Marine Drugs 10, no. 6: 1345-1359. https://doi.org/10.3390/md10061345



