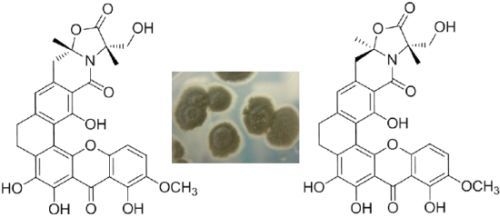
Four New Antibacterial Xanthones from the Marine-Derived Actinomycetes Streptomyces caelestis
Abstract
:1. Introduction
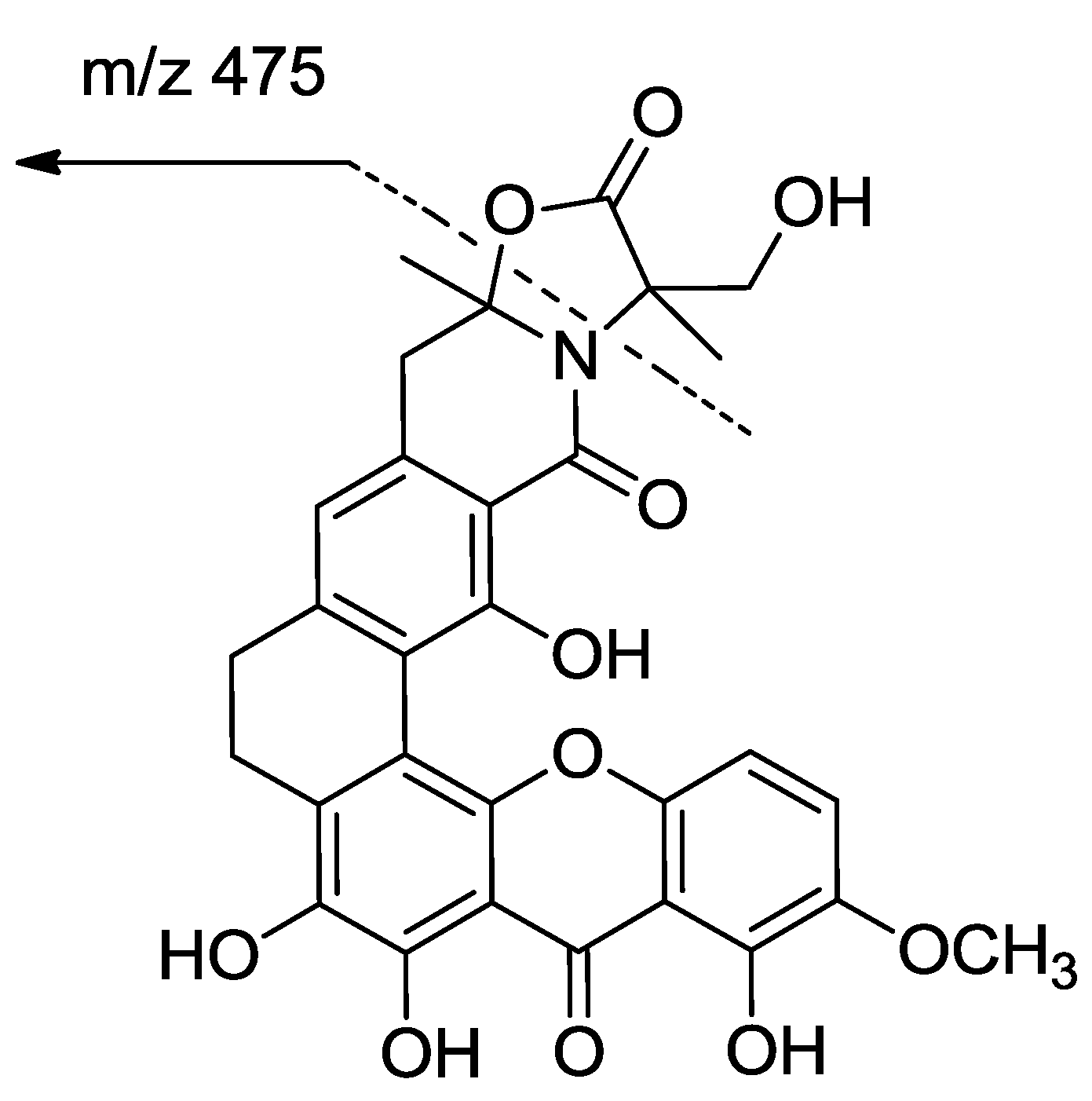
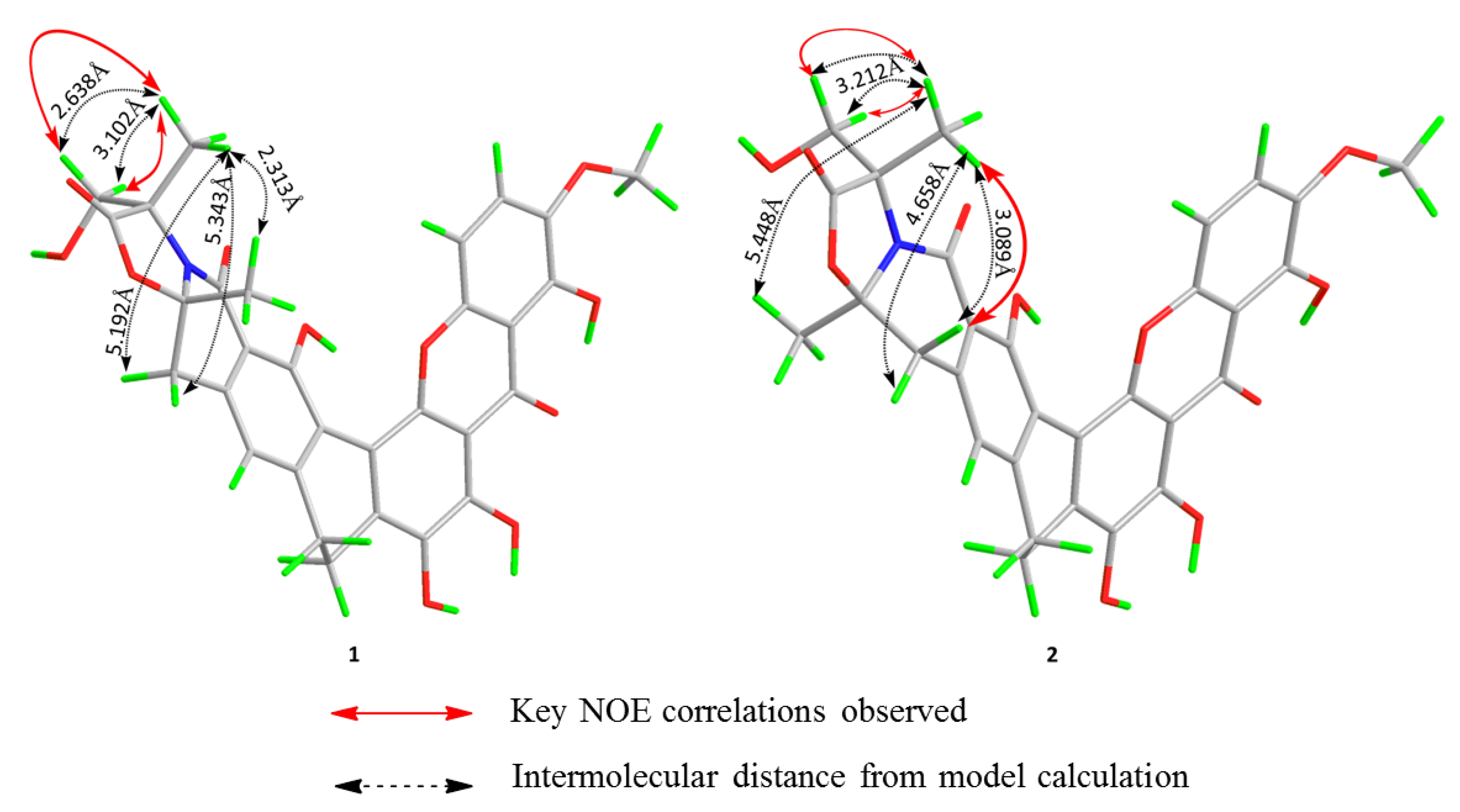
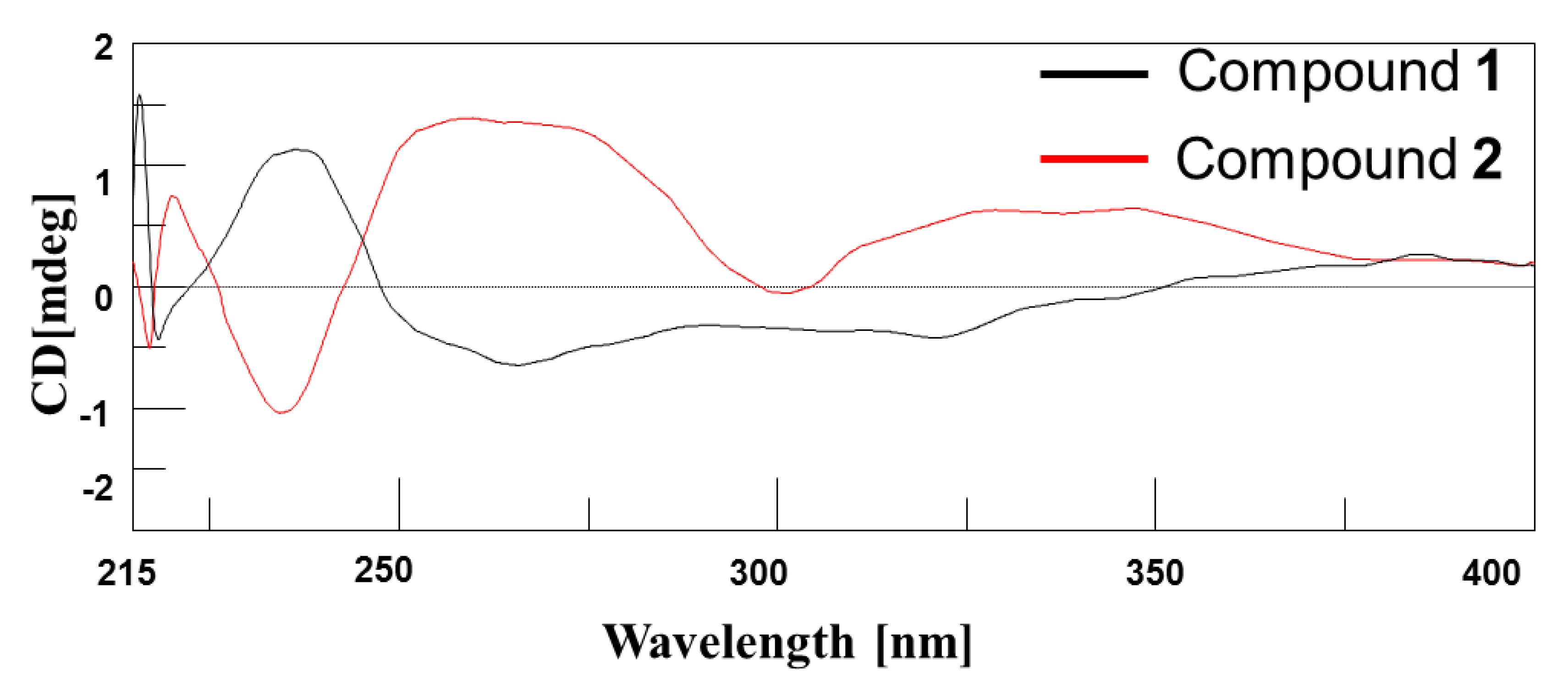
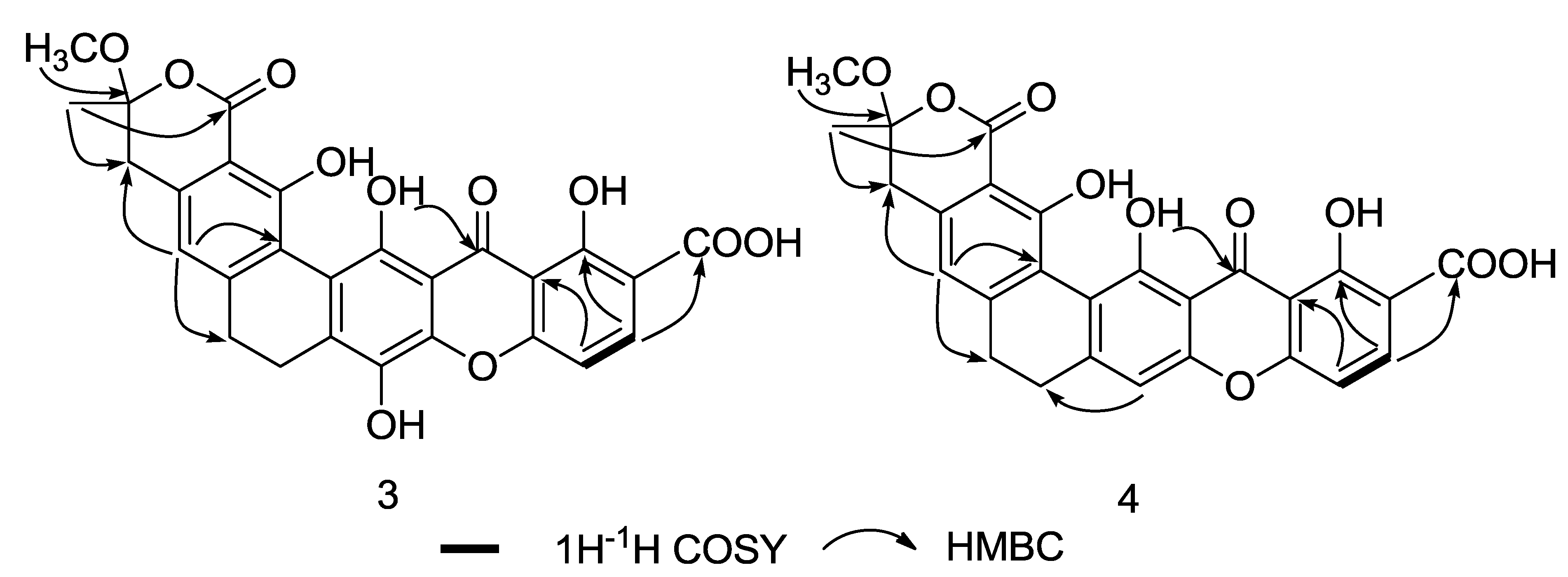
2. Results and Discussion
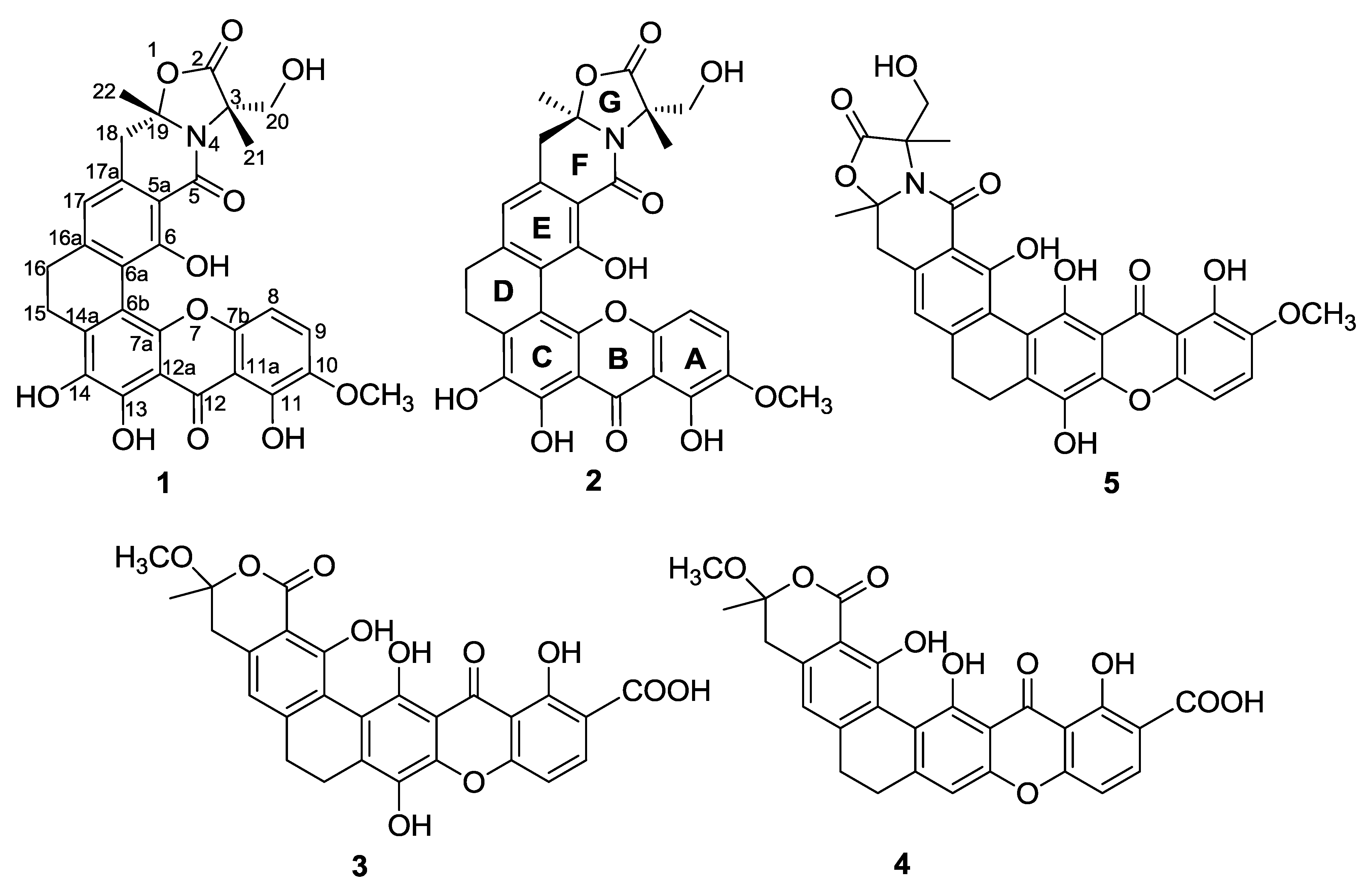
| Position | 1 | 2 | ||
|---|---|---|---|---|
| δC a | δH (J in Hz) b | δC a | δH (J in Hz) b | |
| 2 | 173.7, qC | 173.4, qC | ||
| 3 | 65.5, qC | 65.6, qC | ||
| 5 | 166.3, qC | 166.0, qC | ||
| 5a | 109.6, qC | 109.2, qC | ||
| 6 | 147.2, qC | 146.9, qC | ||
| 6a | 118.3, qC | 118.1, qC | ||
| 6b | 102.6, qC | 102.1, qC | ||
| 7a | 150.6, qC | 150.2, qC | ||
| 7b | 145.9, qC | 145.5, qC | ||
| 8 | 107.7, CH | 6.87, d (8.8) | 107.2, CH | 6.82, d (8.9) |
| 9 | 122.6, CH | 7.57, d (8.8) | 122.3, CH | 7.54, d (8.9) |
| 10 | 149.5, qC | 149.1, qC | ||
| 11 | 143.7, qC | 143.3, qC | ||
| 11a | 106.4, qC | 105.9, qC | ||
| 12 | 183.1, qC | 182.7, qC | ||
| 12a | 108.3, qC | 107.9, qC | ||
| 13 | 159.2, qC | 158.7, qC | ||
| 14 | 137.3, qC | 136.7, qC | ||
| 14a | 134.6, qC | 134.9, qC | ||
| 15 | 23.6, CH2 | 4.09, m | 23.2, CH2 | 4.05, m |
| 16 | 29.4, CH2 | 3.72, m | 29.6, CH2 | 3.69, m |
| 16a | 142.3, qC | 142.0, qC | ||
| 17 | 118.2, CH | 6.90, s | 117.8, CH | 6.87, s |
| 17a | 141.8, qC | 141.4, qC | ||
| 18 | 40.4, CH2 | 3.37, m | 40.0, CH2 | 3.45, m |
| 19 | 93.6, qC | 93.7, qC | ||
| 20 | 61.6, CH2 | 3.55, d (10.9) | 63.4, CH2 | 3.69, d (11.1) |
| 4.31, d (10.9) | 4.10, d (11.1) | |||
| 21 | 19.2, CH3 | 1.61, s | 18.6, CH3 | 1.61, s |
| 22 | 26.5, CH3 | 1.67, s | 25.0, CH3 | 1.69, s |
| -OCH3 | 57.3, CH3 | 3.85, s | 56.9, CH3 | 3.82, s |
| 6-OH | 12.01, s | 12.01, s | ||
| 11-OH | 11.77, s | 11.75, s | ||
| 14-OH | 9.23, s | 9.23, s | ||
| Position | 3 | 4 | ||
|---|---|---|---|---|
| δC a | δH (J in Hz) b | δC a | δH (J in Hz) b | |
| 2 | 169.0, qC | 169.2, qC | ||
| 2a | 106.6, qC | 106.7, qC | ||
| 3 | 158.4, qC | 158.0, qC | ||
| 3a | 118.3, qC | 118.8, qC | ||
| 3b | 112.5, qC | 110.5, qC | ||
| 4 | 150.2, qC | 149.9, qC | ||
| 4a | 106.6, qC | 106.7, qC | ||
| 5 | 180.9, qC | 181.2, qC | ||
| 5a | 117.4, qC | 117.9, qC | ||
| 6 | 150.1, qC | 149.9, qC | ||
| 7 | 116.7, qC | 115.9, qC | ||
| 8 | 125.1, CH | 7.49, d (9.1) | 125.6, CH | 7.45, d (9.1) |
| 9 | 119.3, CH | 7.63, d (9.1) | 119.3, CH | 7.37, d (9.1) |
| 9a | 148.6, qC | 147.1, qC | ||
| 10a | 144.1, qC | 144.9, qC | ||
| 11 | 137.5, qC | 118.7, qC | 6.90, s | |
| 11a | 132.3, qC | 135.9, qC | ||
| 12 | 23.1, CH2 | 2.24, m | 23.0, CH2 | 2.23, m |
| 3.31, overlap | 3.25, m | |||
| 13 | 29.6, CH2 | 2.63, m | 29.2, CH2 | 2.64, m |
| 13a | 148.8, qC | 148.9, qC | ||
| 14 | 117.5, CH | 6.82, s | 117.9, CH | 6.84, s |
| 14a | 137.4, qC | 137.7, qC | ||
| 15 | 36.8, CH2 | 3.16, m | 37.8, CH2 | 3.22, m |
| 3.38, m | 3.41, m | |||
| 16 | 106.1, qC | 106.4, qC | ||
| 17 | 21.7, CH3H | 1.66, s | 22.4, CH3 | 1.69, s |
| 18 | 167.5, qC | 167.6, qC | ||
| –OCH3 | 50.1 | 3.31, s | 50.1 | 3.35, s |
| 3-OH | 11.7, s | 11.8, s | ||
| 4-OH | 12.6, s | 12.8, s | ||
| 11-OH | 9.29, s | 9.22, s | ||
| 18-OH | 10.3, s | 10.3, s | ||
| Compound | Antibacterial (MIC, μg/mL) | Cytotoxicity (IC50μg/mL) | |||
|---|---|---|---|---|---|
| Staphylococcus haemolyticus UST950701-004 | Staphylococcus aureus UST950701-005 | Bacillus subtillis 769 | Staphylococcus aureus ATCC43300 | HeLa cells | |
| 1 | 0.5 | 1.0 | 0.25 | 0.25 | 0.055 |
| 2 | 0.5 | 1.0 | 0.25 | 0.25 | 0.072 |
| 3 | 8.0 | 16.0 | 8.0 | 8.0 | >40 |
| 4 | 8.0 | 16.0 | 8.0 | NA | >40 |
| Penicillin G | 0.13 | 0.25 | 0.13 | NA | NT |
| Streptomycin | 8.0 | 8.0 | 8.0 | NA | NT |
| Cisplatin | NT | NT | NT | NT | 18.14 |
3. Experimental Section
3.1. General Experimental Procedures
3.2. Sample Collection and Microbial Material
3.3. Fermentation, Extraction, and Isolation
3.4. Antibacterial Assays
3.5. Cytotoxic Assays
4. Conclusions
Acknowledgments
Supplementary Files
References and Notes
- Gerwick, W.; Moore, B. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar]
- Bhakuni, D.S.; Rawat, D.S. Bioactive Marine Natural Products; Springer: New Delhi, India, 2005; pp. 13–17. [Google Scholar]
- Seybold, U.; Kourbatova, E.V.; Johnson, J.G.; Halvosa, S.J.; Wang, Y.F.; King, M.D.; Ray, S.M.; Blumberg, H.M. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care associated blood stream infections. Clin. Infect. Dis. 2006, 42, 647–656. [Google Scholar] [CrossRef]
- Cosgrove, S.E.; Sakoulas, G.; Perencevich, E.N.; Schwaber, M.J.; Karchmer, A.W.; Carmeli, Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: A meta-analysis. Clin. Infect. Dis. 2003, 36, 53–59. [Google Scholar] [CrossRef]
- Engemann, J.J.; Carmeli, Y.; Cosgrove, S.E.; Fowler, V.G.; Bronstein, M.Z.; Trivette, S.L.; Briggs, J.P.; Sexton, D.J.; Kaye, K.S. Adverse clinical and economic outcomes attributable to methicillin resistance among patients with Staphylococcus aureus surgical site infection. Clin. Infect. Dis. 2003, 36, 592–598. [Google Scholar] [CrossRef]
- Cosgrove, S.E.; Qi, Y.; Kaye, K.S.; Harbarth, S.; Karchmer, A.W.; Carmeli, Y. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: Mortality, length of stay, and hospital charges. Infect. Control Hosp. Epidemiol. 2005, 2, 166–174. [Google Scholar]
- Klevens, R.M.; Morrison, M.A.; Nadle, J.; Petit, S.; Gershman, K.; Ray, S.; Harrison, L.H.; Lynfield, R.; Dumyati, G.; Townes, M.; et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 2007, 298, 1763–1771. [Google Scholar]
- Deresinski, S. Methicillin-Resistant Staphylococcus aureus: An evolutionary, epidemiologic, and therapeutic odyssey. Clin. Infect. Dis. 2005, 40, 562–573. [Google Scholar] [CrossRef]
- Leeb, M. Antibiotics: A shot in the arm. Nature 2004, 431, 892–893. [Google Scholar]
- Maiese, W.M.; Lechevalier, M.P.; Lechevalier, H.A.; Korshalla, J.; Goodman, J.; Wildey, M.J.; Kuck, N.; Greenstein, M. LL-E19085 alpha, a novel antibiotic from Micromonospora citrea: Taxonomy, fermentation and biological activity. J. Antibiot. 1989, 42, 846–851. [Google Scholar] [CrossRef]
- Carter, G.T.; Nietsche, J.A.; Williams, D.R.; Borders, D.B. Citreamicins, novel antibiotics from Micromonospora citrea: Isolation, characterization, and structure determination. J. Antibiot. 1990, 43, 504–512. [Google Scholar] [CrossRef]
- Reich, M.F.; Lee, V.J.; Ashcroft, J.; Morton, G.O. An unusual cyclization product from the reaction of citreamicin η with sulfene. J. Org. Chem. 1993, 58, 5288–5290. [Google Scholar]
- Hopp, D.C.; Milanowski, D.J.; Rhea, J.; Jacobsen, D.; Rabenstein, J.; Smith, C.; Romari, K.; Clarke, M.; Francis, L.; Irigoyen, M.; et al. Citreamicins with potent gram-positive activity. J. Nat. Prod. 2008, 71, 2032–2035. [Google Scholar]
- Peoples, A.J.; Zhang, Q.; Millett, W.P.; Rothfeder, M.T.; Pescatore, B.C.; Madden, A.A.; Ling, L.L.; Moore, C.M. Neocitreamicins I and II, novel antibiotics with activity against methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci. J. Antibiot. 2009, 61, 457–463. [Google Scholar]
- Han, Z.; Xu, Y.; McConnell, O.; Liu, L.; Li, Y.; Qi, S.; Huang, X.; Qian, P. Two antimycin A analogues from marine-derived actinomycete Streptomyces lusitanus. Mar. Drugs 2012, 10, 668–676. [Google Scholar] [CrossRef]
- Li, S.; Tian, X.; Niu, S.; Zhang, W.; Chen, Y.; Zhang, H.; Yang, X.; Zhang, W.; Li, W.; Zhang, S.; Ju, J.; Zhang, C. Pseudonocardians A–C, new diazaanthraquinone derivatives from a deep-sea Actinomycete Pseudonocardia sp. SCSIO 01299. Mar. Drugs 2011, 9, 1428–1439. [Google Scholar] [CrossRef]
- Huang, H.; Yang, T.; Ren, X.; Liu, J.; Song, Y.; Sun, A.; Ma, J.; Wang, B.; Zhang, Y.; Huang, C.; Zhang, C.; Ju, J. Cytotoxic angucycline class glycosides from the deep sea actinomycete Streptomyces lusitanus SCSIO LR32. J. Nat. Prod. 2012, 75, 202–208. [Google Scholar] [CrossRef]
- Schleissner, C.; Perez, M.; Losada, A.; Rodriguez, P.; Crespo, C.; Zuniga, P.; Fernandez, R.; Reyes, F.; De la Calle, F. Antitumor actinopyranones produced by Streptomyces albus POR-04-15-053 isolated from a marine sediment. J. Nat. Prod. 2011, 74, 1590–1596. [Google Scholar] [CrossRef]
- Kondratyuk, T.P.; Park, E.J.; Yu, R.; Van Breemen, R.B.; Asolkar, R.N.; Murphy, B.T.; Fenical, W.; Pezzuto, J.M. Novel marine phenazines as potential cancer chemopreventive and anti-inflammatory agents. Mar. Drugs 2012, 10, 451–464. [Google Scholar] [CrossRef]
- X-ray crystallogrphic analysis of compound 1: red crystal, 0.05 × 0.06 × 0.28, C30H23NO10, Mr = 557.49, monoclinic, space group P21/c, a = 15.307(3) Å, b = 7.9472(16) Å, c = 24.715(5) Å, β = 97.18(3)°, V = 2983.0(10) Å3, Z = 4, dx = 1.241 g/cm3, F(000) = 1160, μ(Cu Kα) = 0.795 mm−1. The data collection was performed on a Bruker SMART APEX-II using graphite-monochromated radiation (λ = 1.54178 Å), with 4797 unique reflections collected to θmax = 66.98°, where 2116 reflections were observed [F2 > 2σ(F2)]. The structures were solved by direct methods (SHELXTL, 2008) and refined by full-matrix least-squares on F2. The final R = 0.2224, Rw = 0.5021, and S = 1.848. The final model was not very accurate because of the small size of the crystal. The coordinates of the five-member ring and its side chain could not be determined very precisely.
- Schneider, H.J.; Hacket, F.; Rüdiger, V.; Ikeda, H. NMR studies of cyclodextrins and cyclodextrin complexes. Chem. Rev. 1998, 98, 1755–1786. [Google Scholar] [CrossRef]
- Caccamese, S.; Manna, L.; Scivoli, G. Chiral HPLC separation and CD spectra of the C-2 diastereomers of naringin in grapefruit during maturation. Chirality 2003, 15, 661–667. [Google Scholar] [CrossRef]
- Person, R.V.; Monde, K.; Humpf, H.; Berova, N.; Nakanishi, K. A new approach in exciton-coupled circular dichroism (ECCD)—insertion of an auxiliary stereogenic center. Chirality 1995, 7, 128–135. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar]
- Bozdogan, B.; Appelbaum, P.C. Oxazolidinones: Activity, mode of action, and mechanism of resistance. Int. J. Antimicrob. Agents 2004, 23, 113–119. [Google Scholar] [CrossRef]
- Patel, U.; Yan, Y.P.; Hobbs, F.W., Jr.; Kaczmarczyk, J.; Slee, A.M.; Pompliano, D.L.; Kurilla, M.G.; Bobkova, E.V. Oxazolidinones mechanism of action: Inhibition of the first peptide bond formation. J. Biol. Chem. 2001, 276, 37199–37205. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989; pp. 914–923. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Schmitz, F.J.; Fluit, A.C.; Lückefahr, M.; Engler, B.; Hofmann, B.; Verhoef, J.; Heinz, H.P.; Hadding, U.; Jones, M.E. The effect of reserpine, an inhibitor of multidrug efflux pumps, on the in vitro activities of ciprofloxacin, sparfloxacin and moxifloxacin against clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 1998, 42, 807–810. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Y.; Liu, L.; Han, Z.; Lai, P.Y.; Guo, X.; Zhang, X.; Lin, W.; Qian, P.Y. Five new amicoumacins isolated from a marine-derived bacterium Bacillus subtilis. Mar. Drugs 2012, 10, 319–328. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, L.-L.; Xu, Y.; Han, Z.; Li, Y.-X.; Lu, L.; Lai, P.-Y.; Zhong, J.-L.; Guo, X.-R.; Zhang, X.-X.; Qian, P.-Y. Four New Antibacterial Xanthones from the Marine-Derived Actinomycetes Streptomyces caelestis. Mar. Drugs 2012, 10, 2571-2583. https://doi.org/10.3390/md10112571
Liu L-L, Xu Y, Han Z, Li Y-X, Lu L, Lai P-Y, Zhong J-L, Guo X-R, Zhang X-X, Qian P-Y. Four New Antibacterial Xanthones from the Marine-Derived Actinomycetes Streptomyces caelestis. Marine Drugs. 2012; 10(11):2571-2583. https://doi.org/10.3390/md10112571
Chicago/Turabian StyleLiu, Ling-Li, Ying Xu, Zhuang Han, Yong-Xin Li, Liang Lu, Pok-Yui Lai, Jia-Liang Zhong, Xian-Rong Guo, Xi-Xiang Zhang, and Pei-Yuan Qian. 2012. "Four New Antibacterial Xanthones from the Marine-Derived Actinomycetes Streptomyces caelestis" Marine Drugs 10, no. 11: 2571-2583. https://doi.org/10.3390/md10112571