The Role of Rv1476 in Regulating Stress Response and Intracellular Survival of Mycobacterium tuberculosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cell Culture
2.2. Construction of Rv1476 Overexpression Strains
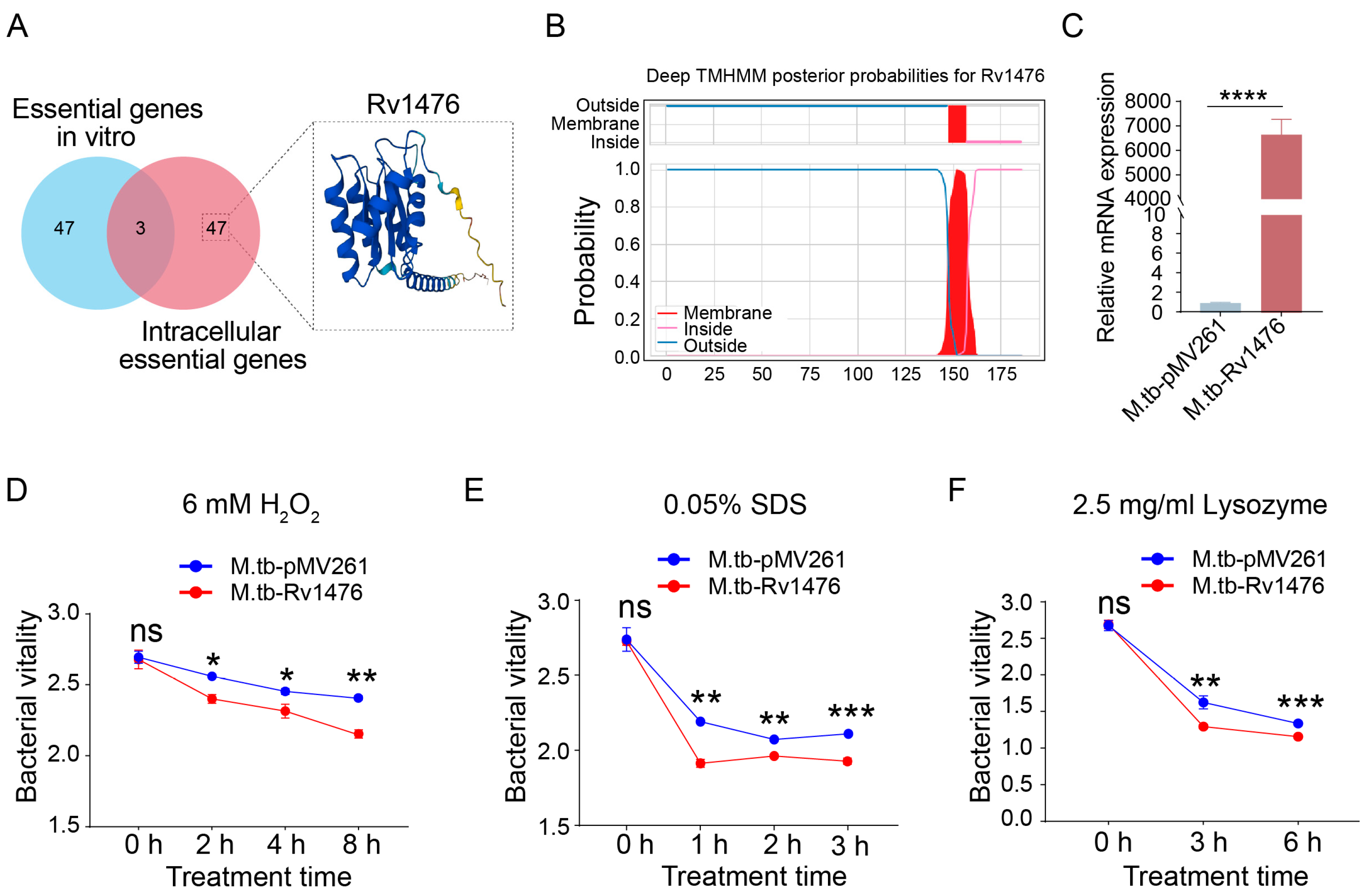
2.3. Survival Analysis under Different Stress Conditions
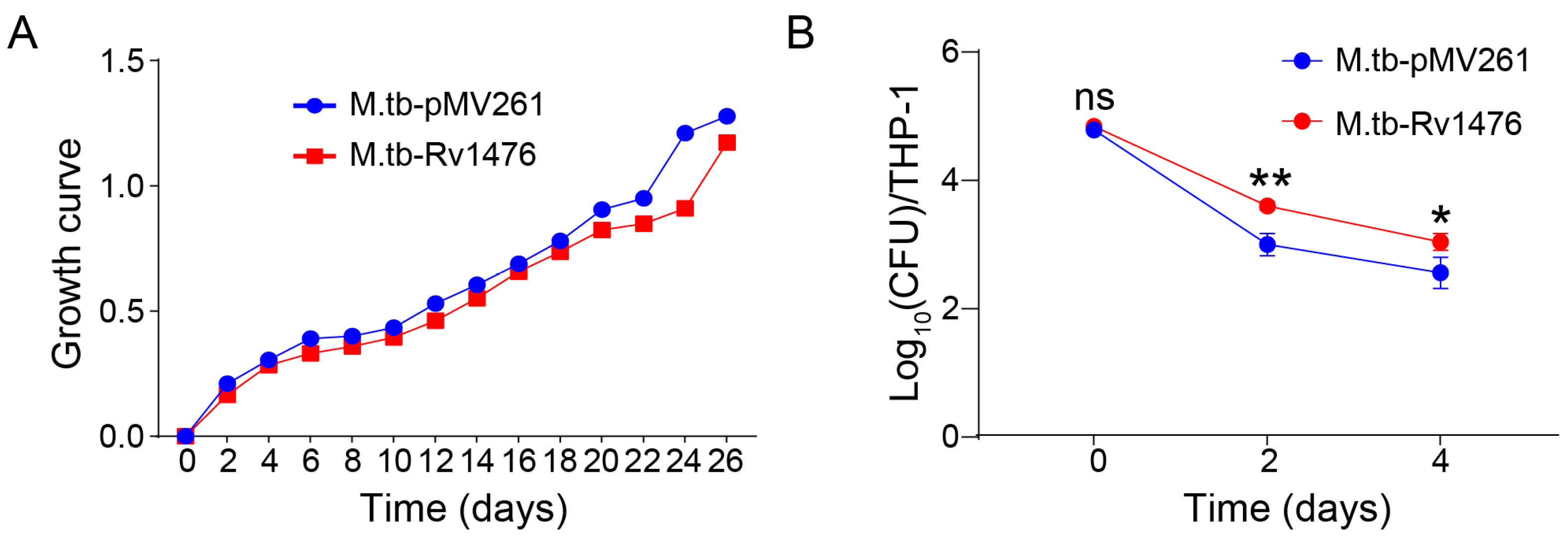
2.4. In Vitro Growth and Intracellular Survival
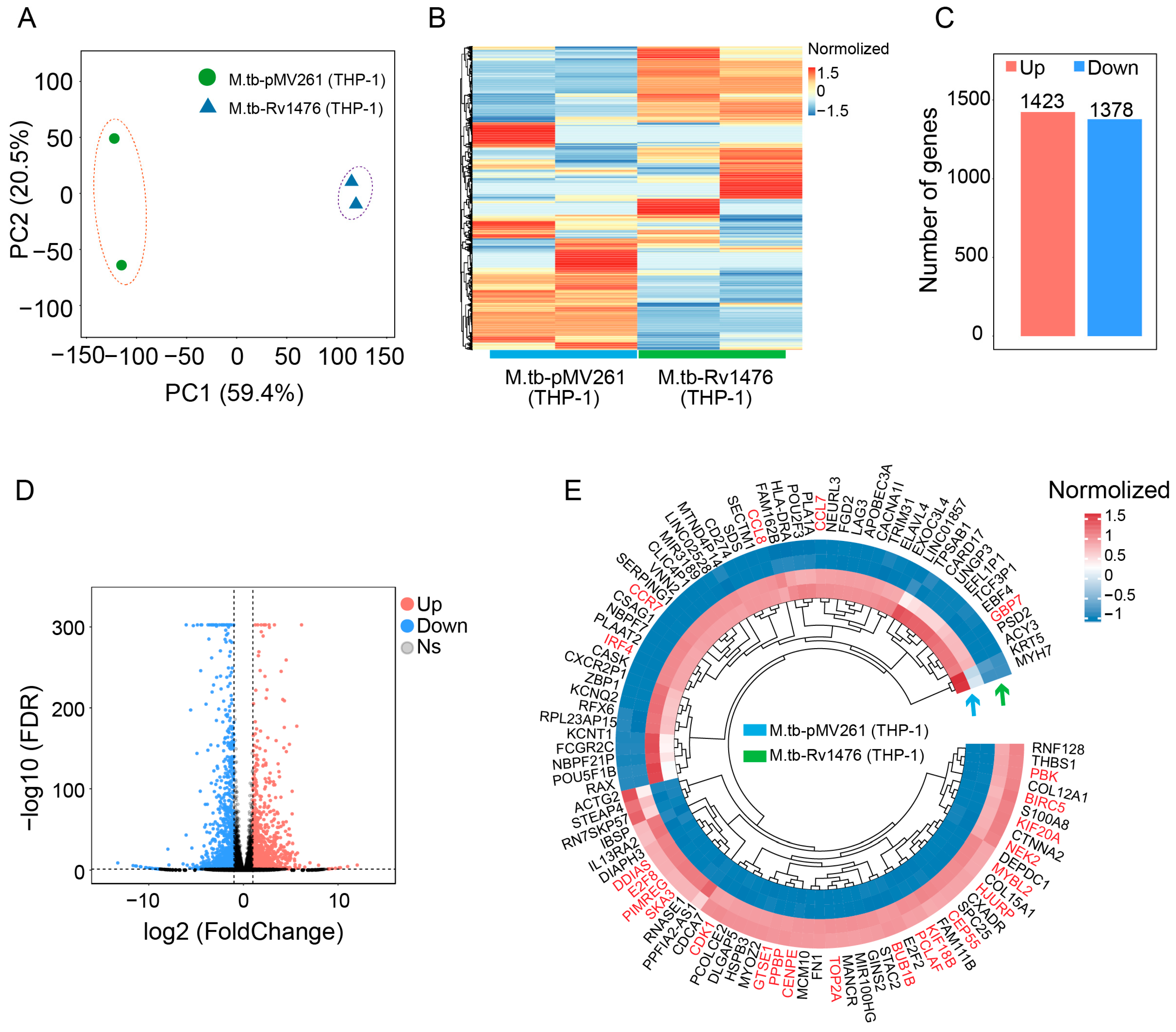
2.5. Host Cell Transcriptome Analysis
2.6. qRT-PCR
2.7. RNA-Seq Data Analysis
2.8. Statistical Analysis
3. Results
3.1. Rv1476 Reduces the Stress Resistance of M. tuberculosis
3.2. Rv1476 Potentiates M. tuberculosis Survival in Macrophage
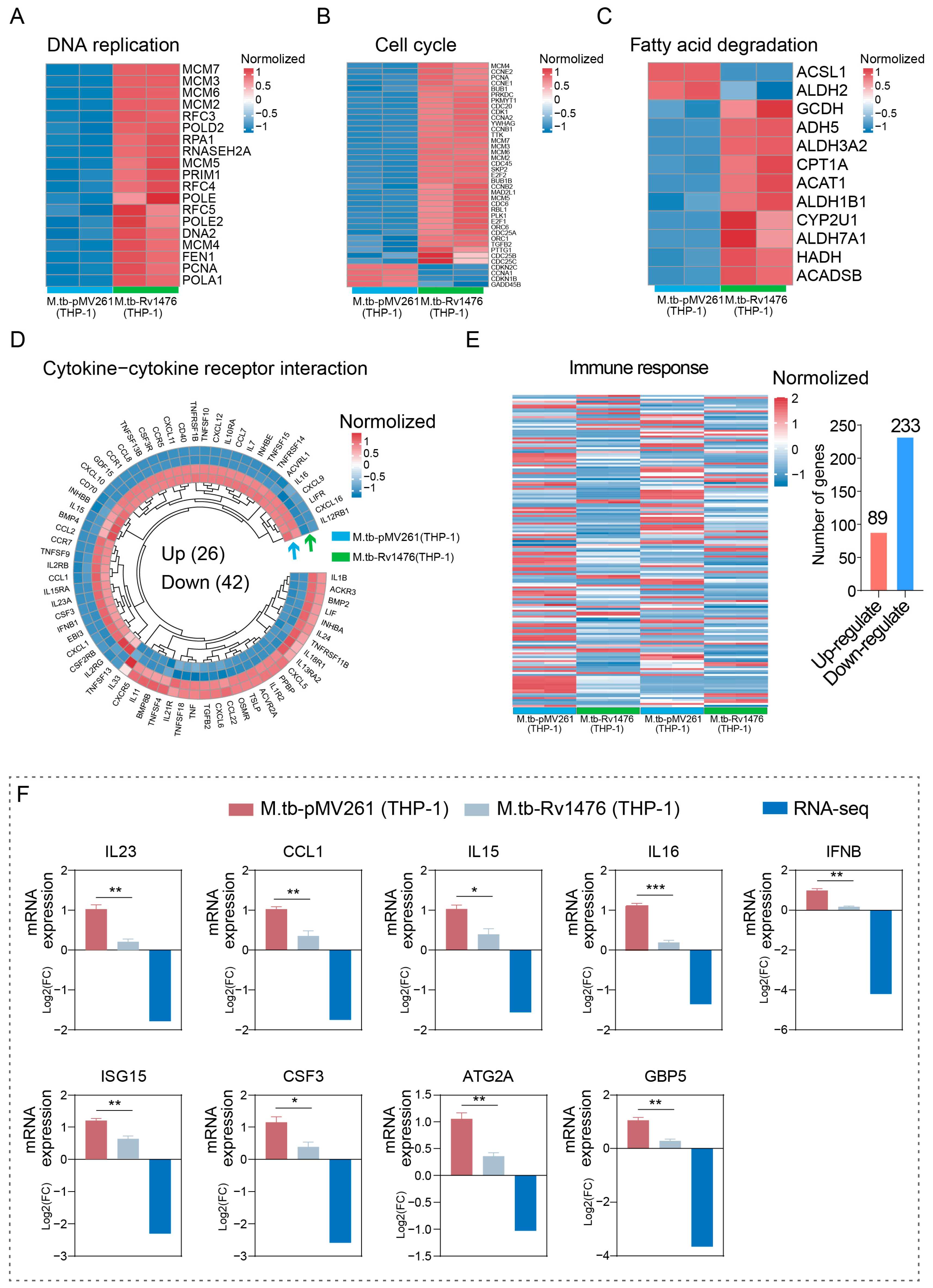
3.3. Rv1476c Regulates Macrophage Gene Expression Profile
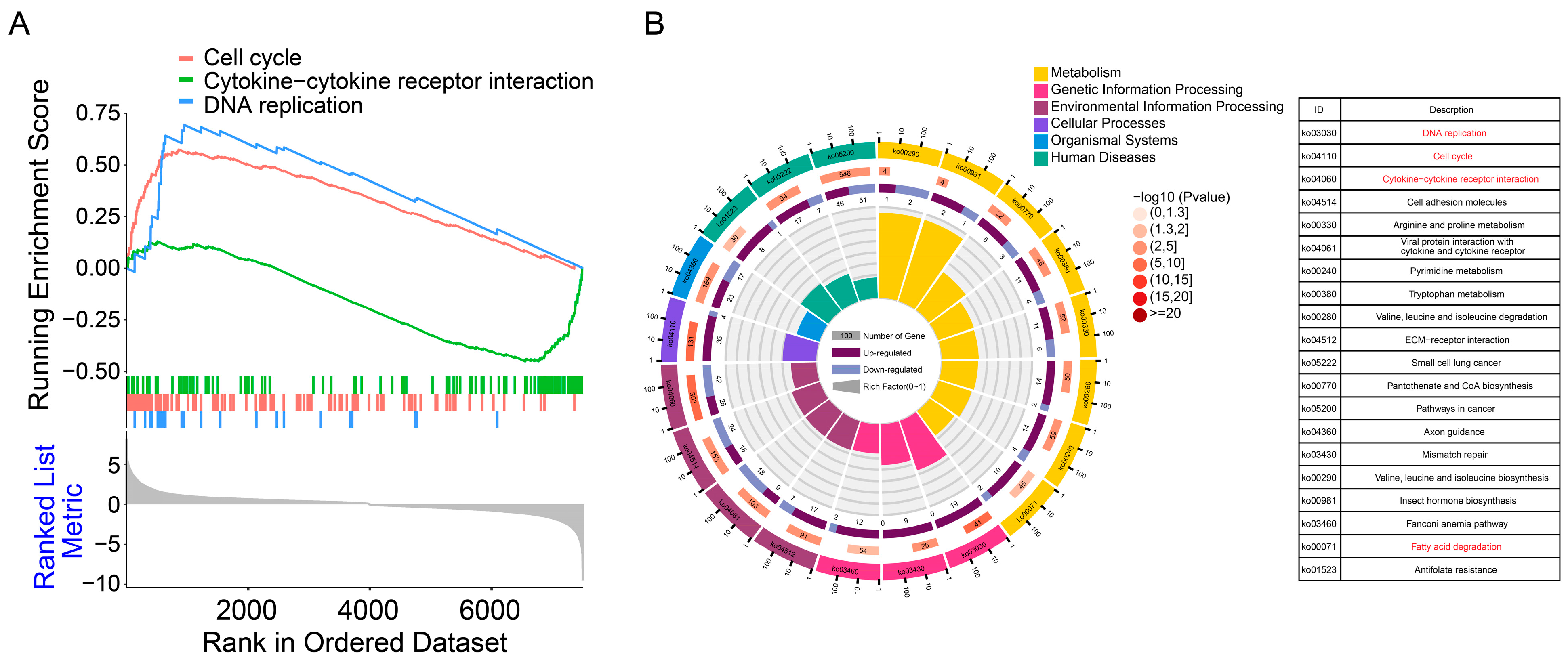
3.4. Pathway Enrichment Analysis of Differentially Expressed Genes
3.5. Rv1476 Affects the Expression of Anti-Tuberculosis-Related Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gopalaswamy, R.; Shanmugam, S.; Mondal, R.; Subbian, S. Of tuberculosis and non-tuberculous mycobacterial infections—A comparative analysis of epidemiology, diagnosis and treatment. J. Biomed. Sci. 2020, 27, 74. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, C.; Xu, Y. Long Non-coding RNAs in Tuberculosis: From Immunity to Biomarkers. Front. Microbiol. 2022, 13, 883513. [Google Scholar] [CrossRef] [PubMed]
- Kramarska, E.; Squeglia, F.; De Maio, F.; Delogu, G.; Berisio, R. PE_PGRS33, an Important Virulence Factor of Mycobacterium tuberculosis and Potential Target of Host Humoral Immune Response. Cells 2021, 10, 161. [Google Scholar] [CrossRef] [PubMed]
- Barry, C.E.; Boshoff, H.I.; Dartois, V.; Dick, T.; Ehrt, S.; Flynn, J.; Schnappinger, D.; Wilkinson, R.J.; Young, D. The spectrum of latent tuberculosis: Rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 2009, 7, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Pieters, J. Mycobacterium tuberculosis and the macrophage: Maintaining a balance. Cell Host Microbe 2008, 3, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Andreu, N.; Phelan, J.; de Sessions, P.F.; Cliff, J.M.; Clark, T.G.; Hibberd, M.L. Primary macrophages and J774 cells respond differently to infection with Mycobacterium tuberculosis. Sci. Rep. 2017, 7, 42225. [Google Scholar] [CrossRef] [PubMed]
- BoseDasgupta, S.; Pieters, J. Macrophage-microbe interaction: Lessons learned from the pathogen Mycobacterium tuberculosis. Semin. Immunopathol. 2018, 40, 577–591. [Google Scholar] [CrossRef]
- Liu, C.H.; Liu, H.; Ge, B. Innate immunity in tuberculosis: Host defense vs pathogen evasion. Cell. Mol. Immunol. 2017, 14, 963–975. [Google Scholar] [CrossRef]
- Weiss, G.; Schaible, U.E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015, 264, 182–203. [Google Scholar] [CrossRef]
- Portevin, D.; Via, L.E.; Eum, S.; Young, D. Natural killer cells are recruited during pulmonary tuberculosis and their ex vivo responses to mycobacteria vary between healthy human donors in association with KIR haplotype. Cell. Microbiol. 2012, 14, 1734–1744. [Google Scholar] [CrossRef]
- Sia, J.K.; Georgieva, M.; Rengarajan, J. Innate Immune Defenses in Human Tuberculosis: An Overview of the Interactions between Mycobacterium tuberculosis and Innate Immune Cells. J. Immunol. Res. 2015, 2015, 747543. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Watts, G.; Crowther, J.E.; Balagopal, A.; Torrelles, J.B.; Robison-Cox, J.; Bargatze, R.F.; Harmsen, A.G.; Crouch, E.C.; Schlesinger, L.S. Mycobacterium tuberculosis binding to human surfactant proteins A and D, fibronectin, and small airway epithelial cells under shear conditions. Infect. Immun. 2006, 74, 3587–3596. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.K.; Sassetti, C.M. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA 2008, 105, 4376–4380. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Chen, J.; Dobos, K.M.; Bradbury, E.M.; Belisle, J.T.; Chen, X. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol. Cell. Proteom. 2003, 2, 1284–1296. [Google Scholar] [CrossRef] [PubMed]
- Malen, H.; Pathak, S.; Softeland, T.; de Souza, G.A.; Wiker, H.G. Definition of novel cell envelope associated proteins in Triton X-114 extracts of Mycobacterium tuberculosis H37Rv. BMC Microbiol. 2010, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- de Souza, G.A.; Leversen, N.A.; Malen, H.; Wiker, H.G. Bacterial proteins with cleaved or uncleaved signal peptides of the general secretory pathway. J. Proteom. 2011, 75, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Minato, Y.; Gohl, D.M.; Thiede, J.M.; Chacon, J.M.; Harcombe, W.R.; Maruyama, F.; Baughn, A.D. Genomewide Assessment of Mycobacterium tuberculosis Conditionally Essential Metabolic Pathways. mSystems 2019, 4, e00070-19. [Google Scholar] [CrossRef]
- Rahman, K.; Jamal, M.; Chen, X.; Zhou, W.; Yang, B.; Zou, Y.; Xu, W.; Lei, Y.; Wu, C.; Cao, X.; et al. Reprogramming Mycobacterium tuberculosis CRISPR System for Gene Editing and Genome-wide RNA Interference Screening. Genom. Proteom. Bioinform. 2022, 20, 1180–1196. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Shim, D.; Kim, H.; Shin, S.J. Mycobacterium tuberculosis Infection-Driven Foamy Macrophages and Their Implications in Tuberculosis Control as Targets for Host-Directed Therapy. Front. Immunol. 2020, 11, 910. [Google Scholar] [CrossRef]
- Mahamed, D.; Boulle, M.; Ganga, Y.; Mc Arthur, C.; Skroch, S.; Oom, L.; Catinas, O.; Pillay, K.; Naicker, M.; Rampersad, S.; et al. Intracellular growth of Mycobacterium tuberculosis after macrophage cell death leads to serial killing of host cells. eLife 2017, 6, e22028. [Google Scholar] [CrossRef]
- Xu, G.; Wang, J.; Gao, G.F.; Liu, C.H. Insights into battles between Mycobacterium tuberculosis and macrophages. Protein Cell. 2014, 5, 728–736. [Google Scholar] [CrossRef]
- Sirakova, T.D.; Thirumala, A.K.; Dubey, V.S.; Sprecher, H.; Kolattukudy, P.E. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 2001, 276, 16833–16839. [Google Scholar] [CrossRef]
- Ghazaei, C. Mycobacterium tuberculosis and lipids: Insights into molecular mechanisms from persistence to virulence. J. Res. Med. Sci. 2018, 23, 63. [Google Scholar] [CrossRef] [PubMed]
- Fieweger, R.A.; Wilburn, K.M.; VanderVen, B.C. Comparing the Metabolic Capabilities of Bacteria in the Mycobacterium tuberculosis Complex. Microorganisms 2019, 7, 177. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.; Li, J.; Niu, J.; Li, P.; Huang, Y.; Li, X.; Duan, W.; Yan, S.; Zhen, J.; Xie, J. Mycobacterium tuberculosis Rv0426c promotes recombinant mycobacteria intracellular survival via manipulating host inflammatory cytokines and suppressing cell apoptosis. Infect. Genet. Evol. 2020, 77, 104070. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, B.X.; Ge, P.P.; Li, J.; Wang, Q.; Gao, G.F.; Qiu, X.B.; Liu, C.H. Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat. Immunol. 2015, 16, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zuo, M.; Chen, H.; Liu, S.; Wu, X.; Cui, Z.; Yang, H.; Liu, H.; Ge, B. Mycobacterium tuberculosis Lipoprotein MPT83 Induces Apoptosis of Infected Macrophages by Activating the TLR2/p38/COX-2 Signaling Pathway. J. Immunol. 2017, 198, 4772–4780. [Google Scholar] [CrossRef] [PubMed]
- Stamm, C.E.; Pasko, B.L.; Chaisavaneeyakorn, S.; Franco, L.H.; Nair, V.R.; Weigele, B.A.; Alto, N.M.; Shiloh, M.U. Screening Mycobacterium tuberculosis Secreted Proteins Identifies Mpt64 as a Eukaryotic Membrane-Binding Bacterial Effector. mSphere 2019, 4, e00354-19. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hartman, M.; Kornfeld, H. Macrophage apoptosis in tuberculosis. Yonsei Med. J. 2009, 50, 1030427. [Google Scholar] [CrossRef] [PubMed]
- Keane, J.; Remold, H.G.; Kornfeld, H. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 2000, 164, 2016–2020. [Google Scholar] [CrossRef] [PubMed]
- Durrbaum-Landmann, I.; Gercken, J.; Flad, H.D.; Ernst, M. Effect of in vitro infection of human monocytes with low numbers of Mycobacterium tuberculosis bacteria on monocyte apoptosis. Infect. Immun. 1996, 64, 5384–5389. [Google Scholar] [CrossRef]
- Dong, W.; Nie, X.; Zhu, H.; Liu, Q.; Shi, K.; You, L.; Zhang, Y.; Fan, H.; Yan, B.; Niu, C.; et al. Mycobacterial fatty acid catabolism is repressed by FdmR to sustain lipogenesis and virulence. Proc. Natl. Acad. Sci. USA 2021, 118, e2019305118. [Google Scholar] [CrossRef]
| Primers | Forward Sequence | Reverse Sequence |
|---|---|---|
| Rv1476-OE | CATCAGGAGGAATCATCTAGA ATGACCGGGCCATATTTTCCT | GTTAACTACGTCGACATCGAT CTAGACGCCTTGATTGAGATC |
| Rv1476-qPCR | ACCATTCCAAGACCGGCAAT | CAGTTGCATCAACCGAGCAC |
| CCL1 | TTGGATGGGTTCAGAGGCAC | GGTAGTTTCGGGGACAGGTG |
| IL15 | AGTTGGCCCAAAGCACCTAA | GCACTGAAACAGCTGCACAA |
| IL16 | GCTGGGATGGAAATGCTCCT | TGTTTGTAGGTGACCGCCTC |
| ISG15 | GTTCATGAATCTGCGCCTGC | AGCCTTTATTTCCGGCCCTT |
| GBP5 | CCGCTGCATACAAATCAGGC | ACATGGGGTCTGACATGTGG |
| IFNB | AGTGTCAGAAGCTCCTGTGG | TAGATGGTCAATGCGGCGTC |
| ATG2A | GAGATCGCCGGCCAGAAG | CAGGTCACGCTGGTTGATGA |
| CSF3 | CTGGTGAGTGAGTGTGCCA | CCGCTATGGAGTTGGCTCAAG |
| IL23A | CCAGCTTCATGCCTCCCTAC | TTGAAGCGGAGAAGGAGACG |
| GAPDH | GACAGTCAGCCGCATCTTCT | GCGCCCAATACGACCAAATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reheman, A.; Wang, Y.; Cai, H.; Wei, P.; Cao, G.; Chen, X. The Role of Rv1476 in Regulating Stress Response and Intracellular Survival of Mycobacterium tuberculosis. Curr. Issues Mol. Biol. 2024, 46, 1556-1566. https://doi.org/10.3390/cimb46020100
Reheman A, Wang Y, Cai H, Wei P, Cao G, Chen X. The Role of Rv1476 in Regulating Stress Response and Intracellular Survival of Mycobacterium tuberculosis. Current Issues in Molecular Biology. 2024; 46(2):1556-1566. https://doi.org/10.3390/cimb46020100
Chicago/Turabian StyleReheman, Aikebaier, Yifan Wang, Huaiyuan Cai, Pingyang Wei, Gang Cao, and Xi Chen. 2024. "The Role of Rv1476 in Regulating Stress Response and Intracellular Survival of Mycobacterium tuberculosis" Current Issues in Molecular Biology 46, no. 2: 1556-1566. https://doi.org/10.3390/cimb46020100






