Effects of Essential Amino Acid Deficiency on General Control Nonderepressible 2/Eukaryotic Initiation Factor 2 Signaling and Proteomic Changes in Primary Bovine Mammary Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Cell Viability
2.3. Cell Proliferation
2.4. Total Protein
2.5. Western Blot
2.6. Proteomic Quantification
2.7. Statistical Analysis
3. Results
3.1. Viability, Proliferation, and Total Protein Concentration of MECs
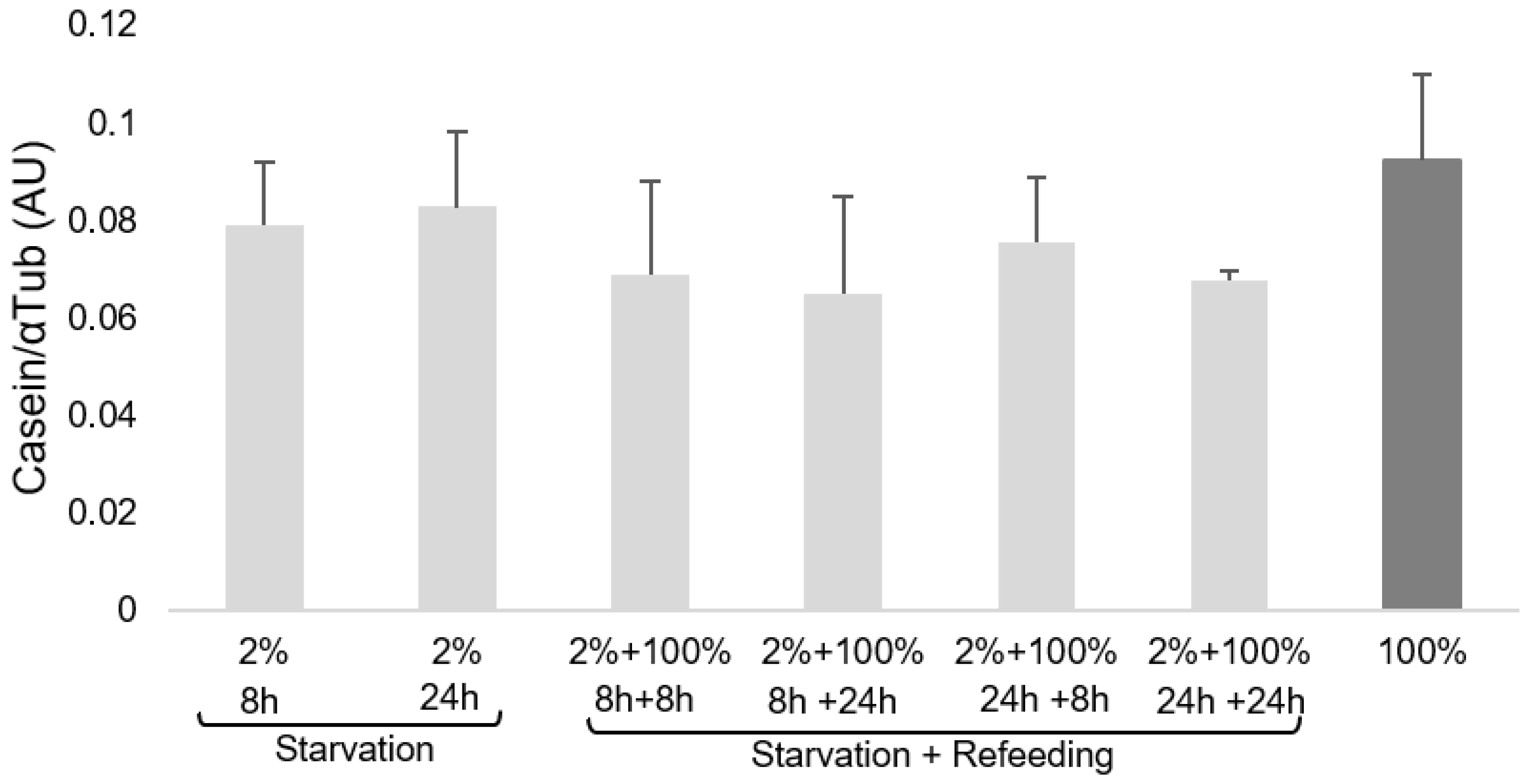
3.2. Casein Expression in MECs
3.3. Protein Expression in the GCN2/eIF2 Pathway
3.4. MEC Proteome under Depletion and Repletion Conditions
3.5. Factors Regulating PS
3.5.1. Responses to EAA Depletion for 8 and 24 h
3.5.2. Responses to Repletion after 8 h of EAA Depletion
3.5.3. Responses to Repletion after 24 h of EAA Depletion
4. Discussion
4.1. Lactocyte (MEC) Model Validity
4.2. Casein and Related Proteins
4.3. Relationship between the GCN2/eIF2 Pathway and MEC Protein Expression (Proteome)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carroll, M.C. Complement and humoral immunity. Vaccine 2008, 26, 128–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edick, A.M.; Audette, J.; Burgos, S.A. CRISPR-Cas9-mediated knockout of GCN2 reveals a critical role in sensing amino acid deprivation in bovine mammary epithelial cells. J. Dairy Sci. 2021, 104, 1123–1135. [Google Scholar] [CrossRef] [PubMed]
- Broer, S.; Broer, A. Amino acid homeostasis and signalling in mammalian cells and organisms. Biochem. J. 2017, 474, 1935–1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connors, M.T.; Poppi, D.P.; Cant, J.P. Protein elongation rates in tissues of growing and adult sheep. J. Anim. Sci. 2008, 86, 2288–2295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appuhamy, J.A.; Knoebel, N.A.; Nayananjalie, W.A.; Escobar, J.; Hanigan, M.D. Isoleucine and leucine independently regulate mTOR signaling and protein synthesis in MAC-T cells and bovine mammary tissue slices. J. Nutr. 2012, 142, 484–491. [Google Scholar] [CrossRef] [Green Version]
- Arriola Apelo, S.I.; Singer, L.M.; Lin, X.; Ray, W.K.; Helm, R.F.; Hanigan, M.D. Mammary gland from lactating cows responded additively to individual essential amino acids in casein synthesis rate. In Energy and Protein Metabolism and Nutrition in Sustainable Animal Production, Proceedings of the 4th International Symposium on Energy and Protein Metabolism and Nutrition, Sacramento, CA, USA, 9–12 September 2013; Oltjen, J.W., Kebreab, E., Lapierre, H., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2013; pp. 449–450. [Google Scholar] [CrossRef]
- Nichols, K.; van Laar, H.; Bannink, A.; Dijkstra, J. Mammary gland utilization of amino acids and energy metabolites differs when dairy cow rations are isoenergetically supplemented with protein and fat. J. Dairy Sci. 2019, 102, 1160–1175. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Jiang, M.; Shi, Y.; Song, S.; Hou, X.; Lin, Y. Prolactin regulates LAT1 expression via STAT5 (signal transducer and activator of transcription 5) signaling in mammary epithelial cells of dairy cows. J. Dairy Sci. 2020, 103, 6627–6634. [Google Scholar] [CrossRef]
- Janjanam, J.; Jamwal, M.; Singh, S.; Kumar, S.; Panigrahi, A.K.; Hariprasad, G.; Jena, M.K.; Anand, V.; Kumar, S.; Kaushik, J.K.; et al. Proteome analysis of functionally differentiated bovine (Bos indicus) mammary epithelial cells isolated from milk. Proteomics 2013, 13, 3189–3204. [Google Scholar] [CrossRef]
- Roncada, P.; Piras, C.; Soggiu, A.; Turk, R.; Urbani, A.; Bonizzi, L. Farm animal milk proteomics. J. Proteom. 2012, 75, 4259–4274. [Google Scholar] [CrossRef] [PubMed]
- Janjanam, J.; Singh, S.; Jena, M.K.; Varshney, N.; Kola, S.; Kumar, S.; Kaushik, J.K.; Grover, S.; Dang, A.K.; Mukesh, M.; et al. Comparative 2D-DIGE proteomic analysis of bovine mammary epithelial cells during lactation reveals protein signatures for lactation persistency and milk yield. PLoS ONE 2014, 9, e102515. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; Ning, C.; Dong, Y.; Zhao, P.; Li, J.; Fan, Z.; Li, J.; Yu, Y.; Mrode, R.; Liu, J.F. Quantitative proteome analysis of bovine mammary gland reveals protein dynamic changes involved in peak and late lactation stages. Biochem. Biophys. Res. Commun. 2017, 494, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, J.; Bu, D.; Wei, H.; Zhou, L.; Li, F.; Loor, J.J. In vitro culture and characterization of a mammary epithelial cell line from Chinese Holstein dairy cow. PLoS ONE 2009, 4, e7636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Xu, B.; Wang, H.; Bu, D.; Wang, J.; Loor, J.J. Effects of Arginine concentration on the in vitro expression of Casein and mTOR pathway related genes in mammary epithelial cells from dairy cattle. PLoS ONE 2014, 9, e95985. [Google Scholar] [CrossRef] [PubMed]
- Arriola Apelo, S.I.; Singer, L.M.; Lin, X.Y.; McGilliard, M.L.; St-Pierre, N.R.; Hanigan, M.D. Isoleucine, leucine, methionine, and threonine effects on mammalian target of rapamycin signaling in mammary tissue. J. Dairy Sci. 2014, 97, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Kuharev, J.; Navarro, P.; Distler, U.; Jahn, O.; Tenzer, S. In-depth evaluation of software tools for data-independent acquisition based label-free quantification. Proteomics 2015, 15, 3140–3151. [Google Scholar] [CrossRef]
- Distler, U.; Kuharev, J.; Navarro, P.; Tenzer, S. Label-free quantification in ion mobility-enhanced data-independent acquisition proteomics. Nat. Protoc. 2016, 11, 795–812. [Google Scholar] [CrossRef]
- RCoreTeam. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 18 October 2018).
- Rose, M.T.; Aso, H.; Yonekura, S.; Komatsu, T.; Hagino, A.; Ozutsumi, K.; Obara, Y. In vitro differentiation of a cloned bovine mammary epithelial cell. J. Dairy Res. 2002, 69, 345–355. [Google Scholar] [CrossRef]
- Castilho, B.A.; Shanmugam, R.; Silva, R.C.; Ramesh, R.; Himme, B.M.; Sattlegger, E. Keeping the eIF2 alpha kinase Gcn2 in check. Biochim. Biophys. Acta 2014, 1843, 1948–1968. [Google Scholar] [CrossRef] [Green Version]
- Bellato, H.M.; Hajj, G.N. Translational control by eIF2alpha in neurons: Beyond the stress response. Cytoskeleton 2016, 73, 551–565. [Google Scholar] [CrossRef]
- Dever, T.E.; Green, R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2012, 4, a013706. [Google Scholar] [CrossRef] [Green Version]
- Dever, T.E.; Dinman, J.D.; Green, R. Translation Elongation and Recoding in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef]
- Malta-Vacas, J.; Aires, C.; Costa, P.; Conde, A.R.; Ramos, S.; Martins, A.P.; Monteiro, C.; Brito, M. Differential expression of the eukaryotic release factor 3 (eRF3/GSPT1) according to gastric cancer histological types. J. Clin. Pathol. 2005, 58, 621–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Ma, N.; Gao, X.; Liu, W.; Jia, J.; Tang, L.; Li, M.; Yang, L.; Li, T.; Yan, L.; et al. Role of GSPT1 and GSPT2 polymorphisms in different outcomes upon Hepatitis B virus infection and prognosis to lamivudine therapy. Biosci. Rep. 2019, 39, BSR20181668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, A.J.; Sharp, J.A.; Lefevre, C.M.; Nicholas, K.R. Uncoupling the mechanisms that facilitate cell survival in hormone-deprived bovine mammary explants. J. Mol. Endocrinol. 2008, 41, 103–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osorio, J.S.; Lohakare, J.; Bionaz, M. Biosynthesis of milk fat, protein, and lactose: Roles of transcriptional and posttranscriptional regulation. Physiol. Genom. 2016, 48, 231–256. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Devgan, G.; Darnell, J.E., Jr.; Bromberg, J.F. Constitutively activated Stat3 protects fibroblasts from serum withdrawal and UV-induced apoptosis and antagonizes the proapoptotic effects of activated Stat1. Proc. Natl. Acad. Sci. USA 2001, 98, 1543–1548. [Google Scholar] [CrossRef] [PubMed]
- Groner, B. Transcription factor regulation in mammary epithelial cells. Domest. Anim. Endocrinol. 2002, 23, 25–32. [Google Scholar] [CrossRef]
- Doelman, J.; Kim, J.J.; Carson, M.; Metcalf, J.A.; Cant, J.P. Branched-chain amino acid and lysine deficiencies exert different effects on mammary translational regulation. J. Dairy Sci. 2015, 98, 7846–7855. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, A.A.; Nones, K.; Roy, N.C.; McNabb, W.C.; Mackenzie, D.S.; Pacheco, D.; McCoard, S. Initiation and elongation steps of mRNA translation are involved in the increase in milk protein yield caused by growth hormone administration during lactation. J. Dairy Sci. 2009, 92, 1889–1899. [Google Scholar] [CrossRef]
- Cant, J.P.; Kim, J.J.M.; Cieslar, S.R.L.; Doelman, J. Symposium review: Amino acid uptake by the mammary glands: Where does the control lie? J. Dairy Sci. 2018, 101, 5655–5666. [Google Scholar] [CrossRef] [Green Version]
- Peuhu, E.; Virtakoivu, R.; Mai, A.; Wärri, A.; Ivaska, J. Epithelial vimentin plays a functional role in mammary gland development. Development 2017, 144, 4103–4113. [Google Scholar] [CrossRef] [Green Version]
- Yoder, P.S.; Ruiz-Cortes, T.; Castro, J.J.; Hanigan, M.D. Effects of varying extracellular amino acid profile on intracellular free amino acid concentrations and cell signaling in primary mammary epithelial cells. J. Dairy Sci. 2019; in press. [Google Scholar] [CrossRef]
- Kapila, N.; Sharma, A.; Kishore, A.; Sodhi, M.; Tripathi, P.K.; Mohanty, A.K.; Mukesh, M. Impact of Heat Stress on Cellular and Transcriptional Adaptation of Mammary Epithelial Cells in Riverine Buffalo (Bubalus Bubalis). PLoS ONE 2016, 11, e0157237. [Google Scholar] [CrossRef] [Green Version]


| Protein Group | Protein/Trt | 2% EAA 8 h | 2% EAA 24 h | 2% EAA 8 + 8 h RF | 2% EAA 8 + 24 h RF | 2% EAA 24 + 8 h RF | 2% EAA 24+ 24 h RF |
|---|---|---|---|---|---|---|---|
| 1. Cell proliferation/cell cycle progression | GSPT1 | ↓ | ↑ | ||||
| CSNK2A1 | ↓ | ||||||
| CDCP42 | |||||||
| 2. Transcription factors | STAT1 | ||||||
| STAT3 | |||||||
| 3. Initiation factors that turn off translation or are turned off by eIF2 | eIF1 | ↑ | ↓ | ↑ | ↓ | ||
| eIF1A | ↓ | ||||||
| eIF2S1 | ↓ | ↑ | ↑ | ↓ | |||
| eIF2S3 | |||||||
| eIF3A | |||||||
| eIF3C | |||||||
| eIF3F | ↓ | ↑ | |||||
| eIF3J | |||||||
| eIF5A | ↓ | ||||||
| eIF6 | ↑ | ↑ | |||||
| 4. Initiation factors that turn on translation | eIF3 | ||||||
| eIF4 | |||||||
| eIF4A1 | ↓ | ↓ | ↓ | ||||
| eIF4A2 | |||||||
| eIF4A3 | |||||||
| 5. Elongation factors | eEF1A2 | ||||||
| eEF1B | ↑ | ↓ | |||||
| eEF1D | ↑ | ↓ | |||||
| eEF1G | ↓ | ||||||
| eEF2 | |||||||
| 6. Posttranslational proteins | PEPD | ↓ | |||||
| PPIA | ↑ | ↓ | |||||
| PPIB | ↑ | ↑ | ↓ | ||||
| CSNK2A1 | ↓ | ||||||
| TPP1 | |||||||
| FKBP3 | ↑ | ↓ | ↓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Cortés, Z.T.; Yoder, P.; Hanigan, M.D. Effects of Essential Amino Acid Deficiency on General Control Nonderepressible 2/Eukaryotic Initiation Factor 2 Signaling and Proteomic Changes in Primary Bovine Mammary Epithelial Cells. Curr. Issues Mol. Biol. 2022, 44, 1075-1086. https://doi.org/10.3390/cimb44030071
Ruiz-Cortés ZT, Yoder P, Hanigan MD. Effects of Essential Amino Acid Deficiency on General Control Nonderepressible 2/Eukaryotic Initiation Factor 2 Signaling and Proteomic Changes in Primary Bovine Mammary Epithelial Cells. Current Issues in Molecular Biology. 2022; 44(3):1075-1086. https://doi.org/10.3390/cimb44030071
Chicago/Turabian StyleRuiz-Cortés, Zulma Tatiana, Peter Yoder, and Mark D. Hanigan. 2022. "Effects of Essential Amino Acid Deficiency on General Control Nonderepressible 2/Eukaryotic Initiation Factor 2 Signaling and Proteomic Changes in Primary Bovine Mammary Epithelial Cells" Current Issues in Molecular Biology 44, no. 3: 1075-1086. https://doi.org/10.3390/cimb44030071






