Metformin Enhances TKI-Afatinib Cytotoxic Effect, Causing Downregulation of Glycolysis, Epithelial–Mesenchymal Transition, and EGFR-Signaling Pathway Activation in Lung Cancer Cells
Abstract
:1. Introduction
2. Results
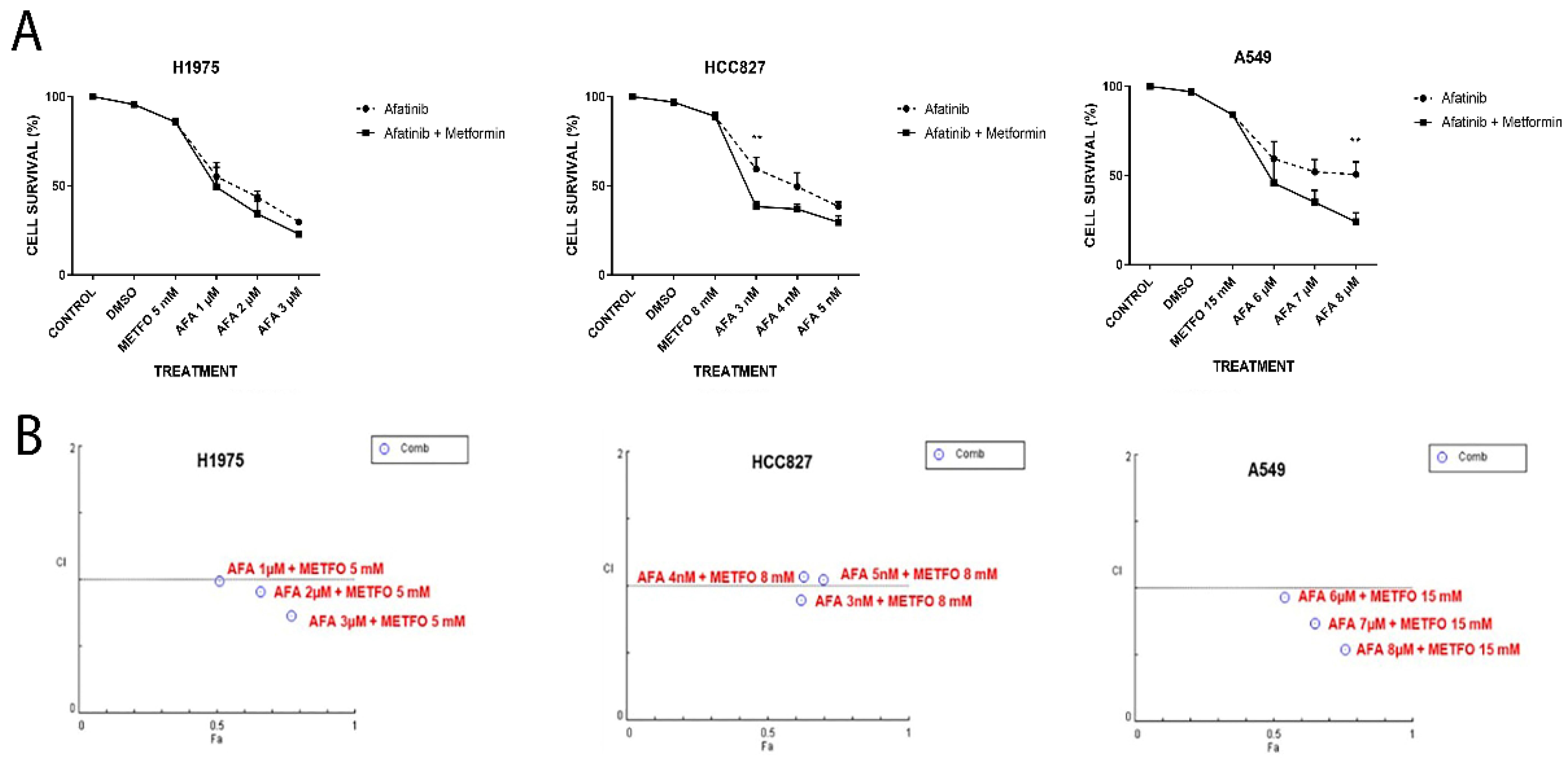
2.1. Unique or Combined (Afatinib and Metformin) Treatments Decrease Cellular Viability of NSCLC Cell Lines in an Additive–Synergistic Manner
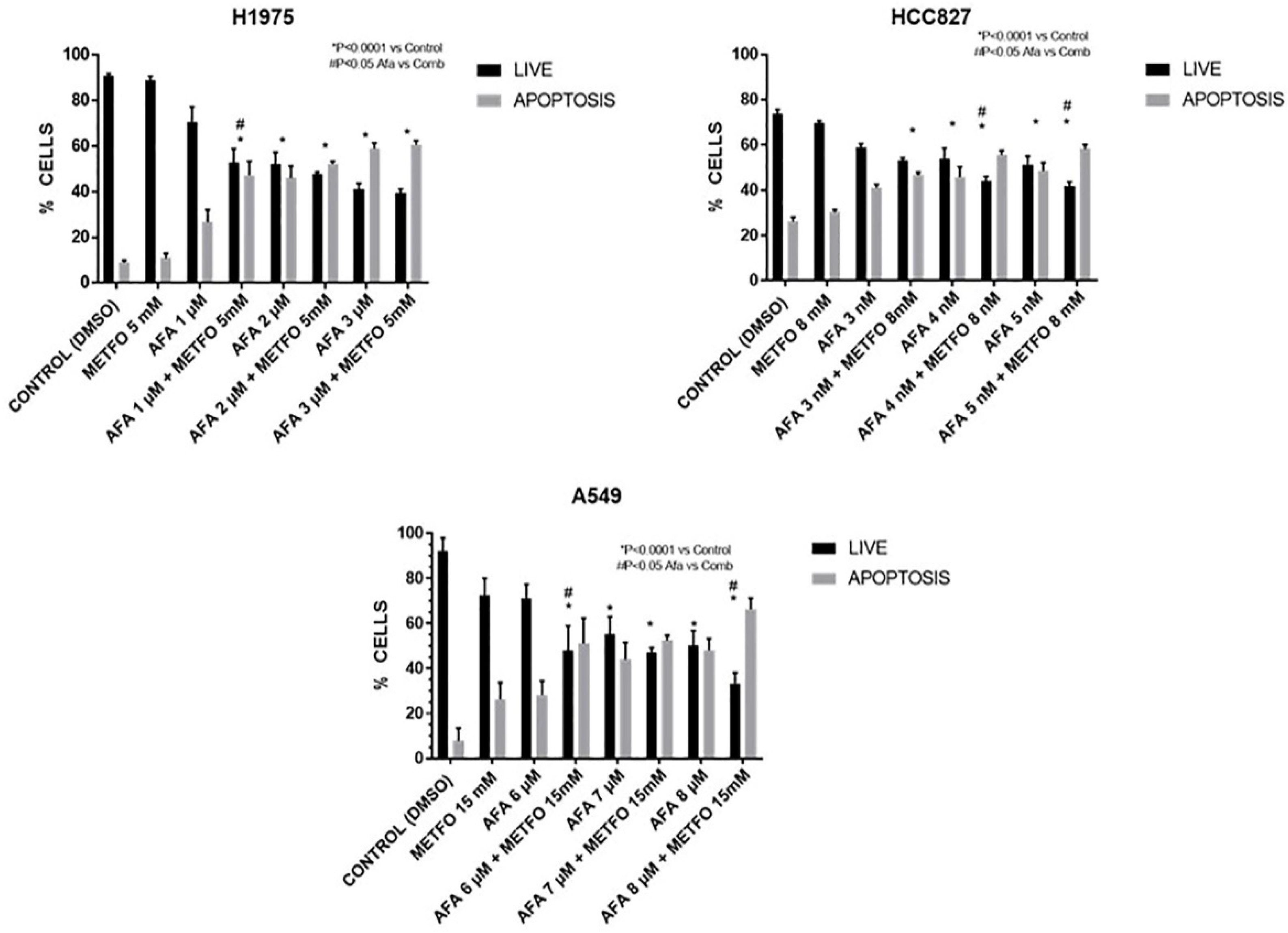
2.2. Combined Treatment of Afatinib Plus Metformin Induces Apoptosis in a Synergic Way
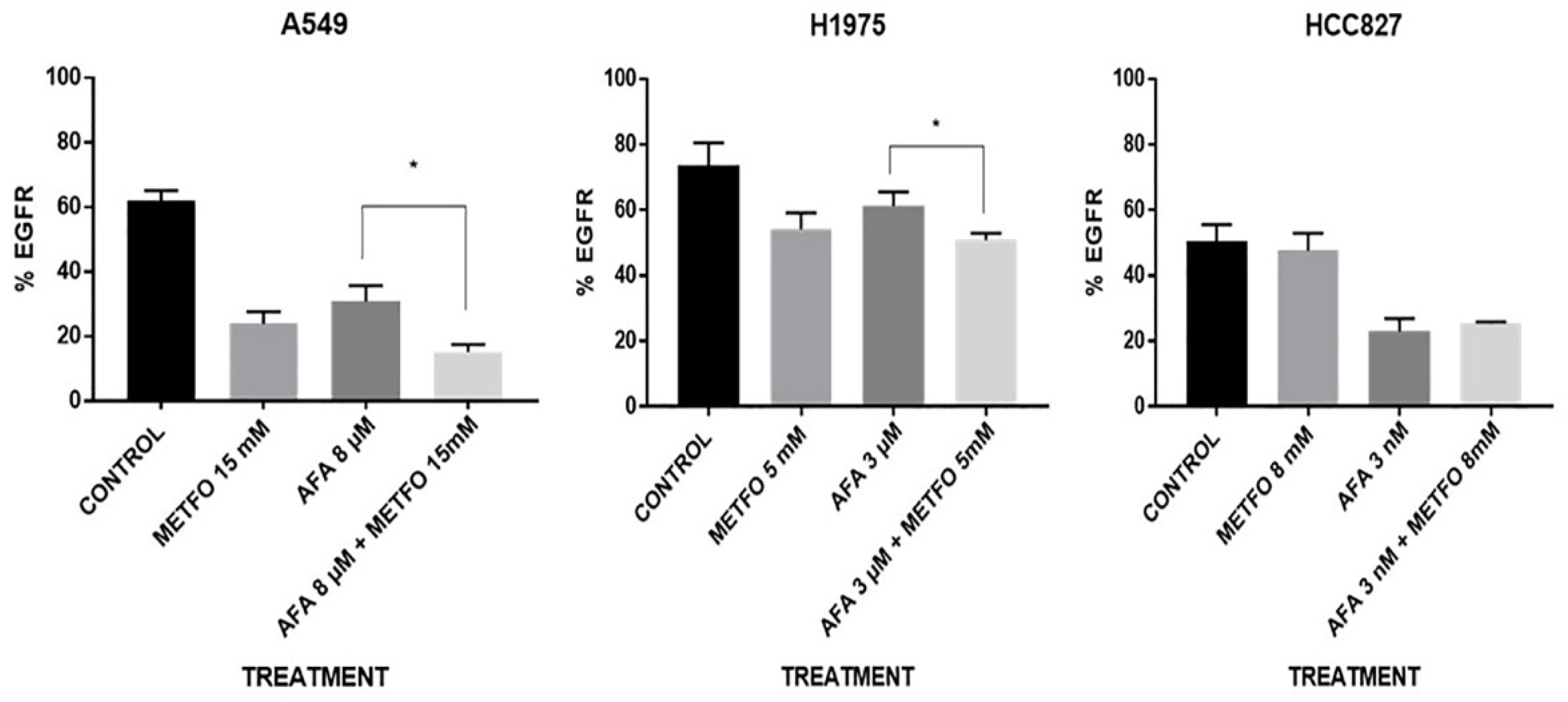
2.3. EGFR Expression at the Cellular Membrane Is Regulated by Combined Afatinib–Metformin Treatment
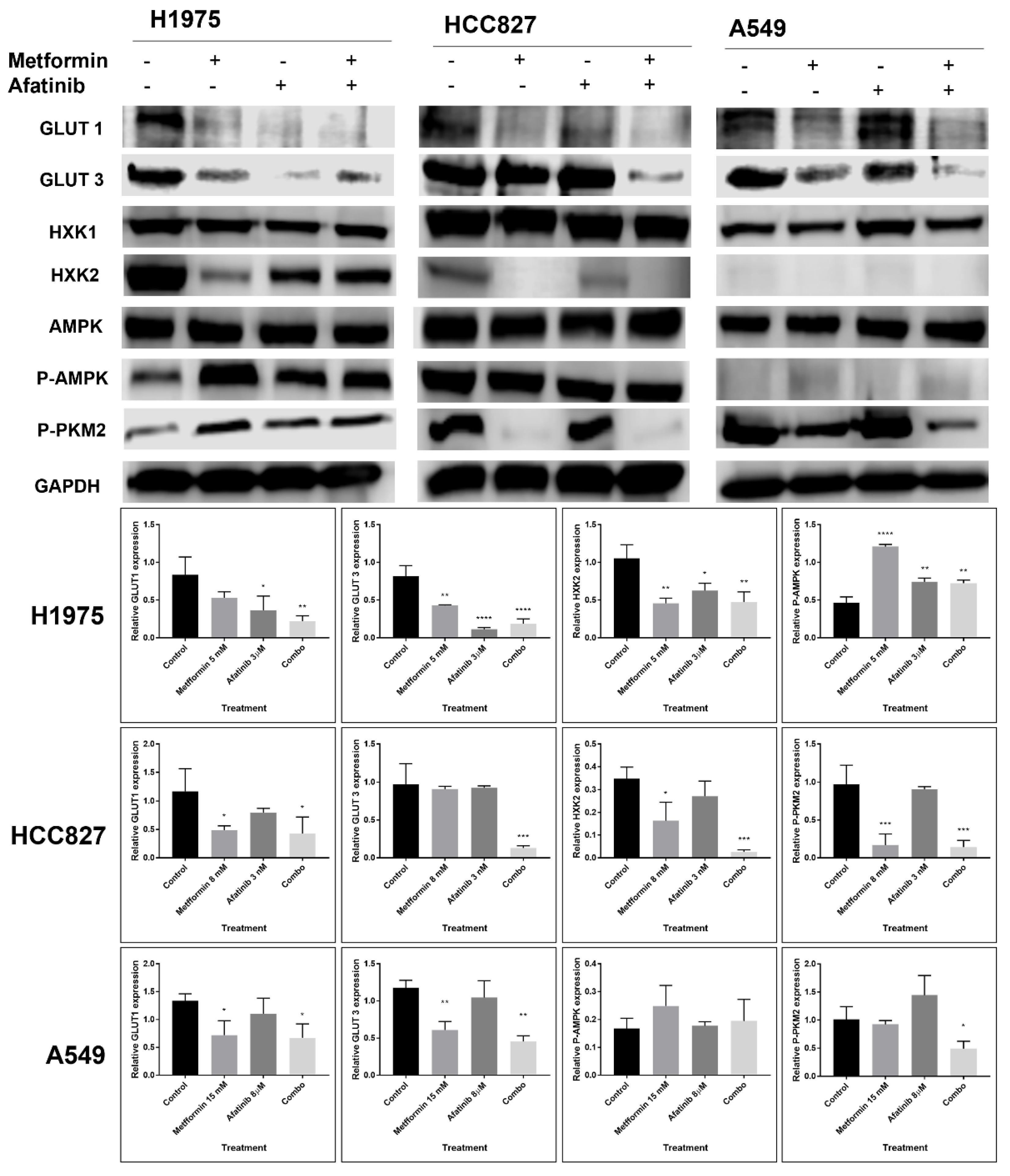
2.4. Metformin Might Sensitize LC Cells to EGFR-TKI Therapy through the Reduction in the Expression and Activation of the EGFR Signaling Pathway
2.5. Combined Metformin/EGFR-TKI Treatment Reduces EMT Biomarkers and Increases Epithelial Marker E-Cadherin in NSCLC Cells
2.6. The Combination Treatment with Metformin–Afatinib Modifies the Glycolytic Phenotype in an EGFR-Mutation Status Dependent Manner
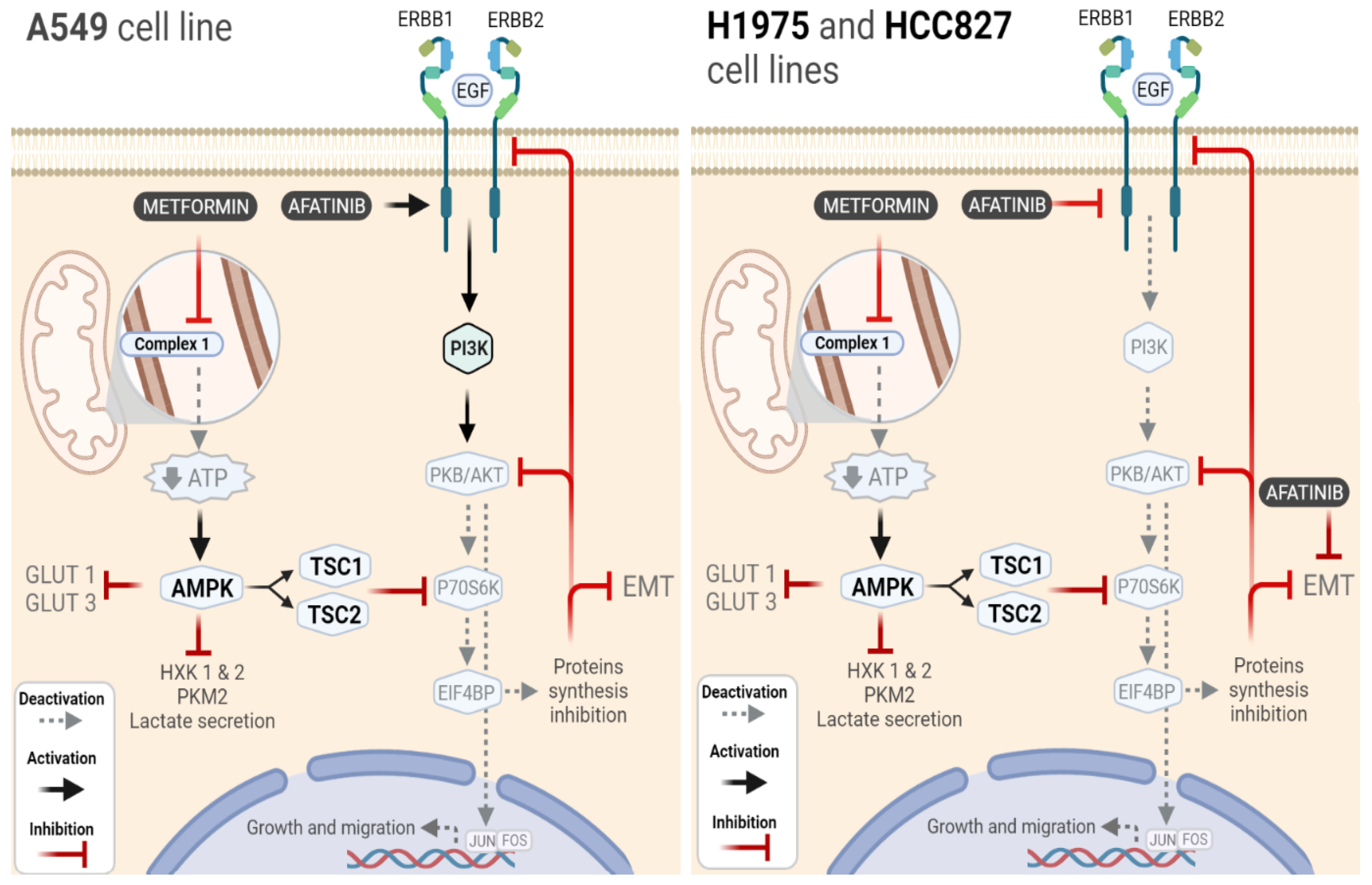
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Treatments
4.2. MTT Assay
4.3. Drug Interaction Analysis
4.4. Apoptosis Assay
4.5. EGFR-Membrane Quantification Assays
4.6. Western Blotting
4.7. Zymogram Assays
4.8. Glucose Uptake Assay
4.9. Lactate Secretion Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barrón-Barrón, F.; Guzmán-De Alba, E.; Alatorre-Alexander, J.; Aldaco-Sarvider, F.; Bautista-Aragón, Y.; Blake-Cerda, M.; Blanco-Vázquez, Y.C.; Campos-Gómez, S.; Corona-Cruz, J.F.; Iñiguez-García, M.A.; et al. National Clinical Practice Guidelines for the management of non-small cell lung cancer in early, locally advanced and metastatic stages. Extended version. Salud Publica Mex. 2019, 61, 359–414. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrot-Zhang, J.; Soca-Chafre, G.; Patterson, N.; Thorner, A.R.; Nag, A.; Watson, J.; Genovese, G.; Rodriguez, J.; Gelbard, M.K.; Corrales-Rodriguez, L.; et al. Genetic ancestry contributes to somatic mutations in lung cancers from admixed latin american populations. Cancer Discov. 2021, 11, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, J.; Malvezzi, M.; Negri, E.; La Vecchia, C.; Boffetta, P. Risk factors for lung cancer worldwide. Eur. Respir. J. 2016, 48, 889–902. [Google Scholar] [CrossRef] [Green Version]
- Arrieta, O.; Cardona, A.F.; Martín, C.; Más-López, L.; Corrales-Rodríguez, L.; Bramuglia, G.; Castillo-Fernandez, O.; Meyerson, M.; Amieva-Rivera, E.; Campos-Parra, A.D.; et al. Updated frequency of EGFR and KRAS mutations in NonSmall-cell lung cancer in Latin America: The Latin-American consortium for the investigation of lung cancer (CLICaP). J. Thorac. Oncol. 2015, 10, 838–843. [Google Scholar] [CrossRef] [Green Version]
- Bethune, G.; Bethune, D.; Ridgway, N.; Xu, Z. Epidermal growth factor receptor (EGFR) in lung cancer: An overview and update. J. Thorac. Dis. 2010, 2, 48. [Google Scholar]
- Rosell, R.; Cardona, A.F.; Arrieta, O.; Aguilar, A.; Ito, M.; Pedraz, C.; Codony-Servat, J.; Santarpia, M. Coregulation of pathways in lung cancer patients with EGFR mutation: Therapeutic opportunities. Br. J. Cancer 2021, 125, 1602–1611. [Google Scholar] [CrossRef]
- Rosell, R.; Karachaliou, N. Co-mutations in EGFR driven non-small cell lung cancer. EBioMedicine 2019, 42, 18–19. [Google Scholar] [CrossRef] [Green Version]
- Morgillo, F.; Della Corte, C.M.; Fasano, M.; Ciardiello, F. Mechanisms of resistance to EGFR-targeted drugs: Lung cancer. ESMO Open 2016, 1, 000060. [Google Scholar] [CrossRef] [Green Version]
- Stewart, E.L.; Tan, S.Z.; Liu, G.; Tsao, M.S. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl. Lung Cancer Res. 2015, 4, 67. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, L.; Liu, L.; Niu, X. EMT-Mediated Acquired EGFR-TKI Resistance in NSCLC: Mechanisms and Strategies. Front. Oncol. 2019, 1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rosa, V.; Iommelli, F.; Monti, M.; Fonti, R.; Votta, G.; Stoppelli, M.P.; Del Vecchio, S. Reversal of warburg effect and reactivation of oxidative phosphorylation by differential inhibition of EGFR signaling pathways in non-small cell lung cancer. Clin. Cancer Res. 2015, 21, 5110–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suda, K.; Tomizawa, K.; Fujii, M.; Murakami, H.; Osada, H.; Maehara, Y.; Yatabe, Y.; Sekido YMitsudomi, T. Epithelial to mesenchymal transition in an epidermal growth factor receptor-mutant lung cancer cell line with acquired resistance to erlotinib. J. Thorac. Oncol. 2011, 6, 1152–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tulchinsky, E.; Demidov, O.; Kriajevska, M.; Barlev, N.A.; Imyanitov, E. EMT: A mechanism for escape from EGFR-targeted therapy in lung cancer. Biochim. Biophys. Acta-Rev. Cancer 2019, 1871, 29–39. [Google Scholar] [CrossRef]
- Xiao, D.; He, J. Epithelial mesenchymal transition and lung cancer. J. Thorac. Dis. 2010, 2, 154. [Google Scholar] [CrossRef]
- Wang, J.; Gao, Q.; Wang, D.; Wang, Z.; Hu, C. Metformin inhibits growth of lung adenocarcinoma cells by inducing apoptosis via the mitochondria-mediated pathway. Oncol. Lett. 2015, 10, 1343–1349. [Google Scholar] [CrossRef] [Green Version]
- Yousef, M.; Tsiani, E. Metformin in lung cancer: Review of In Vitro and In Vivo animal studies. Cancers 2017, 9, 45. [Google Scholar] [CrossRef]
- Evans, J.M.M.; Donnelly, L.A.; Emslie-Smith, A.M.; Alessi, D.R.; Morris, A.D. Metformin and reduced risk of cancer in diabetic patients. Br. Med. J. 2005, 330, 1304–1305. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, P.J.; Ligibel, J.A.; Stambolic, V. Metformin in breast cancer: Time for action. J. Clin. Oncol. 2009, 27, 3271–3273. [Google Scholar] [CrossRef]
- Morgillo, F.; Sasso, F.C.; Della Corte, C.M.; Vitagliano, D.; D’aiuto, E.; Troiani, T.; Martinelli, E.; De Vita, F.; Orditura, M.; De Palma, R.; et al. Synergistic effects of metformin treatment in combination with gefitinib, a selective EGFR tyrosine kinase inhibitor, in LKB1 wild-type NSCLC cell lines. Clin. Cancer Res. 2013, 19, 3508–3519. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Han, R.; Xiao, H.; Lin, C.; Wang, Y.; Liu, H.; Li, K.; Chen, H.; Sun, F.; Yang, Z.; et al. Metformin sensitizes EGFR-TKI-Resistant human lung cancer cells in vitro and in vivo through inhibition of IL-6 signaling and EMT reversal. Clin. Cancer Res. 2014, 20, 2714–2726. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.H.; Jiao, L.; Lin, C.Y.; Lu, C.H.; Li, L.; Chen, H.Y.; Wang YBHe, Y. Combined treatment with metformin and gefitinib overcomes primary resistance to EGFR-TKIs with EGFR mutation via targeting IGF-1R signaling pathway. Biol. Targets Ther. 2018, 12, 75–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foretz, M.; Hébrard, S.; Leclerc, J.; Zarrinpashneh, E.; Soty, M.; Mithieux, G.; Sakamoto, K.; Andreelli, F.; Viollet, B. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J. Clin. Investig. 2010, 120, 2355–2369. [Google Scholar] [CrossRef] [Green Version]
- Gunton, J.E.; Delhanty, P.J.D.; Takahashi, S.I.; Baxter, R.C. Metformin rapidly increases insulin receptor activation in human liver and signals preferentially through insulin-receptor substrate-2. J. Clin. Endocrinol. Metab. 2003, 88, 1323–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pernicova, I.; Korbonits, M. Metformin-Mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 2014, 10, 143–156. [Google Scholar] [CrossRef]
- Arrieta, O.; Barrón, F.; Padilla, M.Á.S.; Avilés-Salas, A.; Ramírez-Tirado, L.A.; Jiménez, M.J.A.; Vergara, E.; Zatarain-Barrón, Z.L.; Hernández-Pedro, N.; Cardona, A.F.; et al. Effect of Metformin Plus Tyrosine Kinase Inhibitors Compared with Tyrosine Kinase Inhibitors Alone in Patients with Epidermal Growth Factor Receptor-Mutated Lung Adenocarcinoma: A Phase 2 Randomized Clinical Trial. JAMA Oncol. 2019, 5, e192553. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jiang, L.; Wang, Y.; Zhao, Y.; Zhang, X.J.; Wu, G.; Zhou, X.; Sun, J.; Bai, J.; Ren, B.; et al. Combination of metformin and gefitinib as first-line therapy for nondiabetic advanced NSCLC patients with EGFR mutations: A randomized, double-blind phase II trial. Clin. Cancer Res. 2019, 25, 6967–6975. [Google Scholar] [CrossRef] [Green Version]
- Han, R.; Jia, Y.; Li, X.; Zhao, C.; Zhao, S.; Liu, S.; Liu, Y.; Chen, D.; Zhang, Q.; Liu, X.; et al. P76.07 Metformin Enhances the Efficacy of EGFR-TKIs in Advanced Non-Small Cell Lung Cancer Patients With Type 2 Diabetes Mellitus. J. Thorac. Oncol. 2021, 16, S588. [Google Scholar] [CrossRef]
- Han, R.; Jia, Y.; Li, X.; Zhao, C.; Zhao, S.; Liu, S.; Liu, Y.; Chen, D.; Zhang, Q.; Liu, X.; et al. Concurrent use of metformin enhances the efficacy of EGFR-TKIs in patients with advanced EGFR-mutant non-small cell lung cancer-an option for overcoming EGFR-TKI resistance. Transl. Lung Cancer Res. 2021, 10, 1277–1291. [Google Scholar] [CrossRef]
- Li Ling Wang, T.; Hu, M.; Zhang, Y.; Chen, H.; Xu, L. Metformin Overcomes Acquired Resistance to EGFR TKIs in EGFR-Mutant Lung Cancer via AMPK/ERK/NF-κB Signaling Pathway. Front. Oncol. 2020, 10, 1605. [Google Scholar] [CrossRef]
- Wang, X.; Chen, K.; Yu, Y.; Xiang, Y.; Kim, J.H.; Gong, W.; Huang, J.; Shi, G.; Li, Q.; Zhou, M.; et al. Metformin sensitizes lung cancer cells to treatment by the tyrosine kinase inhibitor erlotinib. Oncotarget 2017, 8, 109068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashinuma, H.; Takiguchi, Y.; Kitazono, S.; Kitazono-Saitoh, M.; Kitamura, A.; Chiba, T.; Tada, Y.; Kurosu, K.; Sakaida, E.; Sekine, I.; et al. Antiproliferative action of metformin in human lung cancer cell lines. Oncol. Rep. 2012, 28, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, T.T.; Wang, C.Y.; Chen, Y.L.; Lai, C.C.; Chang, F.Y.; Tsai, Y.T.; Chao, C.H.H.; Shiau, C.W.; Huang, Y.C.T.; Yu, C.J.; et al. Afatinib induces apoptosis in NSCLC without EGFR mutation through Elk-1-mediated suppression of CIP2A. Oncotarget 2015, 6, 2164–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, C.T.; Yeh, K.C.; Lee, C.H.; Chen, Y.Y.; Ho, P.J.; Chang, K.Y.; Chen, C.H.; Lai, Y.K.; Chen, C.T. Molecular profiling of afatinib-resistant non-small cell lung cancer cells in vivo derived from mice. Pharmacol. Res. 2020, 161, 105183. [Google Scholar] [CrossRef]
- Liu, Z.; Gao, W. Overcoming acquired resistance of gefitinib in lung cancer cells without T790M by AZD9291 or Twist1 knockdown in vitro and in vivo. Arch. Toxicol. 2019, 93, 1555–1571. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hu, B.; Lv, X.; Chen, S.; Liu, W.; Shao, Z. The Prognostic Role of Ribosomal Protein S6 Kinase 1 Pathway in Patients With Solid Tumors: A Meta-Analysis. Front. Oncol. 2019, 9, 390. [Google Scholar] [CrossRef]
- Guo, Q.; Liu, Z.; Jiang, L.; Liu, M.; Ma, J.; Yang, C.; Han, L.; Nan, K.; Liang, X. Metformin inhibits growth of human non-small cell lung cancer cells via liver kinase B-1-independent activation of adenosine monophosphate-activated protein kinase. Mol. Med. Rep. 2016, 13, 2590–2596. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.M.; Kwon, O.J.; Hong, Y.K.; Kim, J.H.; Solca, F.; Ha, S.J.; Soo, R.A.; Christensen, J.G.; Lee, J.H.; Cho, B.C. Activation of IL-6R/JAK1/STAT3 signaling induces De Novo resistance to irreversible EGFR inhibitors in non-small cell lung cancer with T790M resistance mutation. Mol. Cancer Ther. 2012, 11, 2254–2264. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.O.; Yang, X.; Duan, S.; Tsai, Y.; Strojny, L.R.; Keng, P.; Chen, Y. IL-6 promotes growth and epithelial-mesenchymal transition of CD133+ cells of non-small cell lung cancer. Oncotarget 2016, 7, 6626. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Cheng, X.; Wang, Y.; Han, R.; Li, L.; Xiang, T.; He, L.; Long, H.; Zhu, B.; He, Y. Metformin inhibits the IL-6-induced epithelial-mesenchymal transition and lung adenocarcinoma growth and metastasis. PLoS ONE 2014, 9, e95884. [Google Scholar] [CrossRef] [Green Version]
- Makinoshima, H.; Takita, M.; Saruwatari, K.; Umemura, S.; Obata, Y.; Ishii, G.; Matsumoto, S.; Sugiyama, E.; Ochiai, A.; Abe, R.; et al. Signaling through the phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) axis is responsible for aerobic glycolysis mediated by glucose transporter in epidermal growth factor receptor (EGFR)-mutated lung adenocarcinoma. Journal of Biological Chemistry 2015, 290, 17495–17504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makinoshima, H.; Takita, M.; Matsumoto, S.; Yagishita, A.; Owada, S.; Esumi, H.; Tsuchihara, K. Epidermal growth factor receptor (EGFR) signaling regulates global metabolic pathways in EGFR-mutated lung adenocarcinoma. J. Biol. Chem. 2014, 289, 20813–20823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrzejewski:, S.; Gravel, S.-P.; Pollak, M.; St-Pierre, J. Metformin directly acts on mitochondria to alter cellular bioenergetics. Cancer Metab. 2014, 2, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luengo, A.; Sullivan, L.B.; Heiden, V.G.V. Understanding the complex-I-ty of metformin action: Limiting mitochondrial respiration to improve cancer therapy. BMC Biol. 2014, 12, 82. [Google Scholar] [CrossRef] [Green Version]
- Chun, S.G.; Liao, Z.; Jeter, M.D.; Chang, J.Y.; Lin, S.H.; Komaki, R.U.; Guerrero, T.M.; Mayo, R.C.; Korah, B.M.; Koshy, S.M.; et al. Metabolic Responses to Metformin in Inoperable Early-stage Non-Small Cell Lung Cancer Treated With Stereotactic Radiotherapy: Results of a Randomized Phase II Clinical Trial. Am. J. Clin. Oncol. 2020, 43, 231–235. [Google Scholar] [CrossRef]
- Li, C.; Xue, Y.; Xi, Y.R.; Xie, K.E. Progress in the application and mechanism of metformin in treating non-small cell lung cancer. Oncol. Lett. 2017, 13, 2873–2880. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; He, C.; Huang, X. Metformin partially reverses the carboplatin-resistance in NSCLC by inhibiting glucose metabolism. Oncotarget 2017, 8, 75206. [Google Scholar] [CrossRef] [Green Version]
- Salani, B.; Marini, C.; Rio, A.D.; Ravera, S.; Massollo, M.; Orengo, A.M.; Amaro, A.; Passalacqua, M.; Maffioli, S.; Pfeffer, U.; et al. Metformin impairs glucose consumption and survival in Calu-1 cells by Direct Inhibition of Hexokinase-II. Sci. Rep. 2013, 3, 2070. [Google Scholar] [CrossRef]
- Luo, L.; Jiang, S.; Huang, D.; Lu, N.; Luo, Z. MLK3 Phophorylates AMPK Independently of LKB1. PLoS ONE 2015, 10, e0123927. [Google Scholar] [CrossRef]
- Ross, F.A.; Hawley, S.A.; Auciello, F.R.; Gowans, G.J.; Atrih, A.; Lamont, D.J.; Hardie, D.G. Mechanisms of Paradoxical Activation of AMPK by the Kinase Inhibitors SU6656 and Sorafenib. Cell Chem. Biol. 2017, 24, 813–824. [Google Scholar] [CrossRef] [Green Version]
- Shaw, R.J. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol. 2009, 196, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Cui, J.; Wang, H.; Medina, R.; Zhang, S.; Zhang, X.; Zhuang, Z.; Lin, Y. Metformin enhances anti-cancer effects of cisplatin in meningioma through AMPK-mTOR signaling pathways. Mol. Ther.-Oncolytics 2021, 20, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Sperl, B.; Ullrich, A.; Knyazev, P. Metformin and salinomycin as the best combination for the eradication of NSCLC monolayer cells and their alveospheres (cancer stem cells) irrespective of EGFR, KRAS, EML4/ALK and LKB1 status. Oncotarget 2014, 5, 12877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Qian, W.; Li, J.; Jiang, Z.; Cheng, L.; Yan, B.; Cao, J.; Sun, L.; Zhou, C.; Lei, M.; et al. Loss of AMPK activation promotes the invasion and metastasis of pancreatic cancer through an HSF1-dependent pathway. Mol. Oncol. 2017, 11, 1475–1492. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Yang, W.; Wu, F.; Wang, C.; Yu, L.; Tang, L.; Qiu, B.; Li, Y.; Guo, L.; Wu, M.; et al. Prognostic Significance of AMPK Activation and Therapeutic Effects of Metformin in Hepatocellular Carcinoma. Clin. Cancer Res. 2013, 19, 5372–5380. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Liu, X.; Fu, H.; Zhou, W.; Zhong, D. 2-Deoxyglucose Suppresses ERK Phosphorylation in LKB1 and Ras Wild-Type Non-Small Cell Lung Cancer Cells. PLoS ONE 2016, 11, e0168793. [Google Scholar] [CrossRef]
- Zahra, K.; Dey, T.; Ashish Mishra, S.P.; Pandey, U. Pyruvate Kinase M2 and Cancer: The Role of PKM2 in Promoting Tumorigenesis. Front. Oncol. 2020, 10, 159. [Google Scholar] [CrossRef] [Green Version]
- Majem, B.; Nadal, E.; Muñoz-Pinedo, C. Exploiting metabolic vulnerabilities of Non small cell lung carcinoma. Semin. Cell Dev. Biol. 2020, 98, 54–62. [Google Scholar] [CrossRef]
- Hirpara, J.; Eu, J.Q.; Tan, J.K.M.; Wong, A.L.; Clement, M.V.; Kong, L.R.; Ohi, N.; Tsunoda, T.; Qu, J.; Goh, B.C.; et al. Metabolic reprogramming of oncogene-addicted cancer cells to OXPHOS as a mechanism of drug resistance. Redox Biol. 2019, 25, 101076. [Google Scholar] [CrossRef]
- Martin, M.J.; Eberlein, C.; Taylor, M.; Ashton, S.; Robinson, D.; Cross, D. Inhibition of oxidative phosphorylation suppresses the development of osimertinib resistance in a preclinical model of EGFR-driven lung adenocarcinoma. Oncotarget 2016, 7, 86313–86325. [Google Scholar] [CrossRef] [Green Version]
- Arrieta, O.; Varela-Santoyo, E.; Soto-Perez-de-Celis, E.; Sánchez-Reyes, R.; De la Torre-Vallejo, M.; Muñiz-Hernández, S.; Cardona, A.F. Metformin use and its effect on survival in diabetic patients with advanced non-small cell lung cancer. BMC Cancer 2016, 16, 633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Yang, X.; Sun, Y.; Li, Y.; Dong, Y.; Shan, D.; Yu, Z. Synergistic effect of metformin and EGFR-TKI in the treatment of non-small cell lung cancer. Transl. Cancer Res. 2020, 9, 372–381. [Google Scholar] [CrossRef] [PubMed]



| Cell Lines | Concentrations of Drugs | |
|---|---|---|
| Metformin | Afatinib | |
| A549 | 15 mM | 6 µM (Combo 1) |
| 7 µM IC50 (Combo 2) | ||
| 8 µM (Combo 3) | ||
| H1975 | 5 mM | 1 µM (Combo 1) |
| 2 µM IC50 (Combo 2) | ||
| 3 µM (Combo 3) | ||
| HCC827 | 8 mM | 3 nM (Combo 1) |
| 4 nM IC50 (Combo 2) | ||
| 5 nM (Combo 3) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrios-Bernal, P.; Hernandez-Pedro, N.; Orozco-Morales, M.; Viedma-Rodríguez, R.; Lucio-Lozada, J.; Avila-Moreno, F.; Cardona, A.F.; Rosell, R.; Arrieta, O. Metformin Enhances TKI-Afatinib Cytotoxic Effect, Causing Downregulation of Glycolysis, Epithelial–Mesenchymal Transition, and EGFR-Signaling Pathway Activation in Lung Cancer Cells. Pharmaceuticals 2022, 15, 381. https://doi.org/10.3390/ph15030381
Barrios-Bernal P, Hernandez-Pedro N, Orozco-Morales M, Viedma-Rodríguez R, Lucio-Lozada J, Avila-Moreno F, Cardona AF, Rosell R, Arrieta O. Metformin Enhances TKI-Afatinib Cytotoxic Effect, Causing Downregulation of Glycolysis, Epithelial–Mesenchymal Transition, and EGFR-Signaling Pathway Activation in Lung Cancer Cells. Pharmaceuticals. 2022; 15(3):381. https://doi.org/10.3390/ph15030381
Chicago/Turabian StyleBarrios-Bernal, Pedro, Norma Hernandez-Pedro, Mario Orozco-Morales, Rubí Viedma-Rodríguez, José Lucio-Lozada, Federico Avila-Moreno, Andrés F. Cardona, Rafael Rosell, and Oscar Arrieta. 2022. "Metformin Enhances TKI-Afatinib Cytotoxic Effect, Causing Downregulation of Glycolysis, Epithelial–Mesenchymal Transition, and EGFR-Signaling Pathway Activation in Lung Cancer Cells" Pharmaceuticals 15, no. 3: 381. https://doi.org/10.3390/ph15030381







