Protein Arrays for Multidrug-resistance in Human Leukemia Cell Determination
Abstract
:Introduction
Experimental
Apparatus
Reagents
Modified glass slide preparation
Procedure
Flow cytometry
Statistical analyses
Results and Discussion
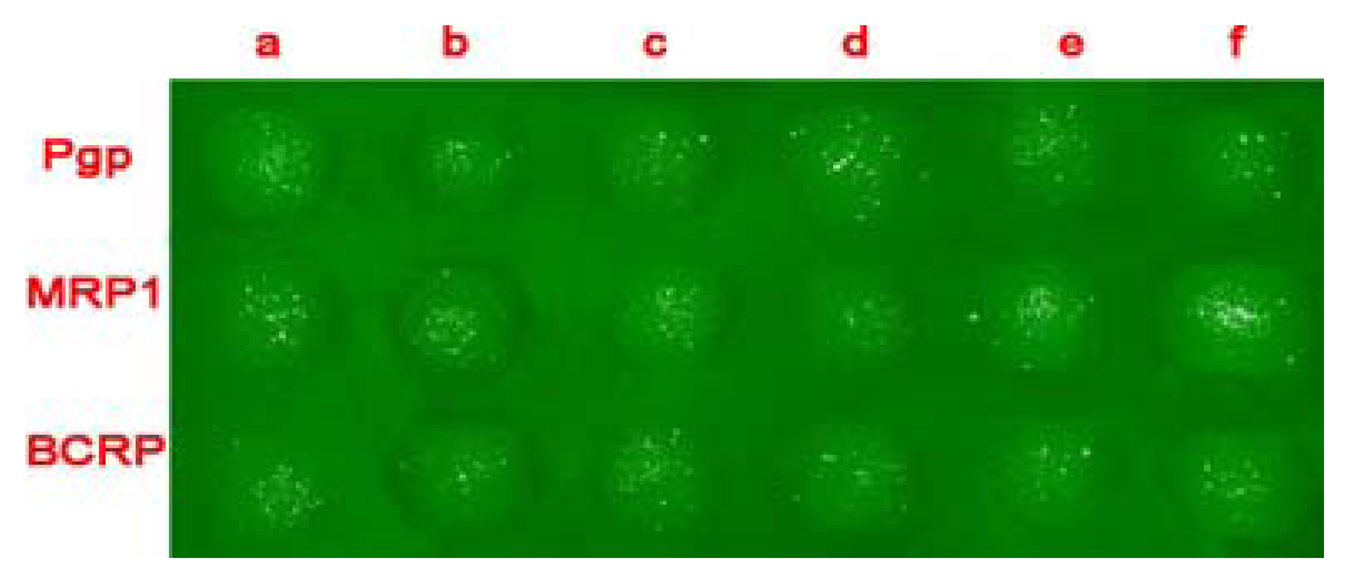
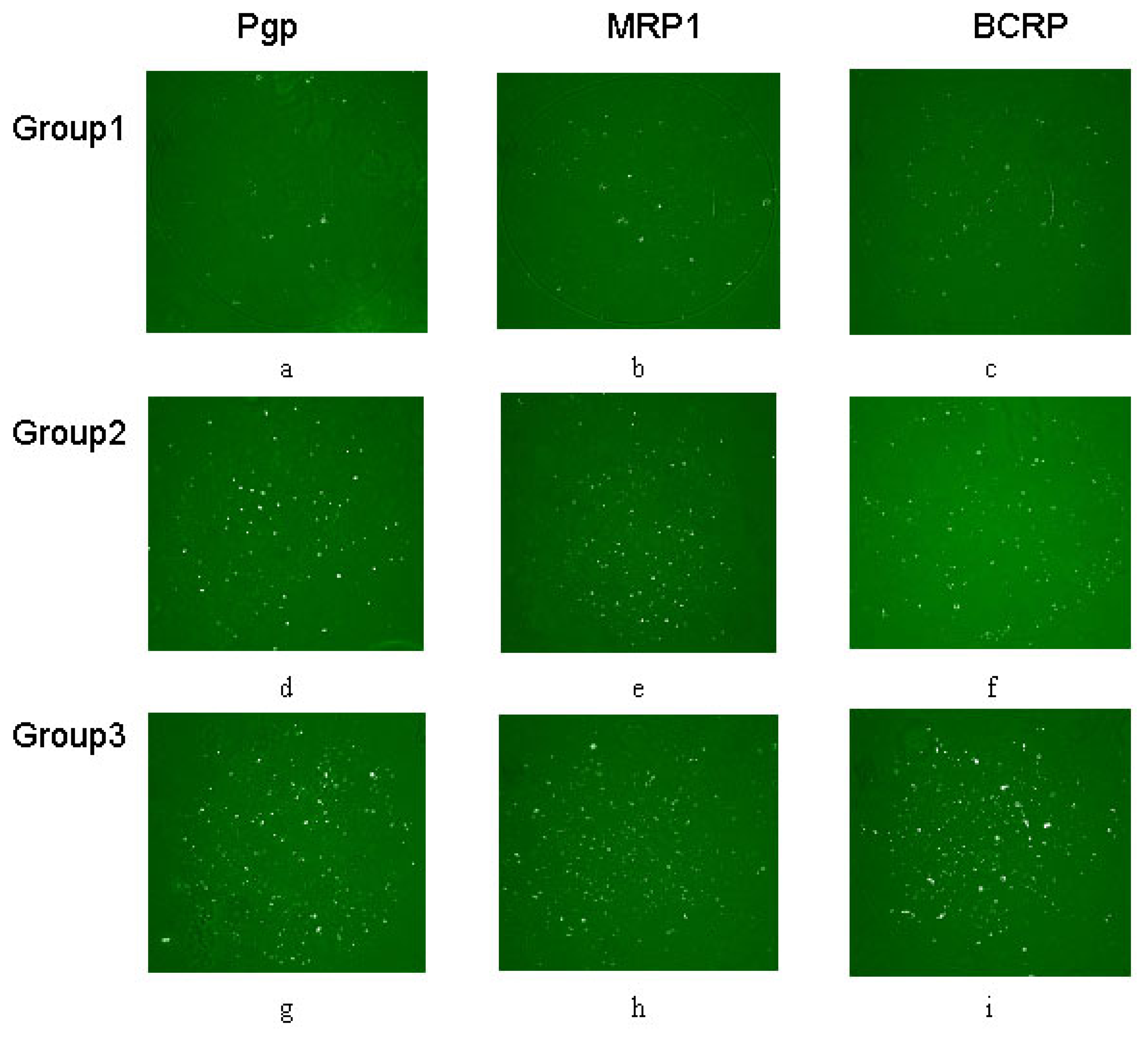
MDR protein arrays for multiplex characterization of leukemia cells autoantibody responses
Cells combine antibody on a spot
Flow cytometry determination results
Conclusions
Acknowledgments
References
- Arceci, R.J. Tumor cell survival and resistance to therapy. Curr. Opin. Hematol. 1996, 3, 279–287. [Google Scholar]
- Moscow, J.A.; Cowan, K.H. Multidrug resistance. J. Natl. Cancer Inst. 1988, 80, 14–20. [Google Scholar]
- Schneider, E.; Hunke, S. ATP-binding–cassette (ABC) transporter systems functional and structural aspects of the ATP-hyfrolyzing subunits/domains. FEMS Microbiol Rev. 1998, 22, 1–20. [Google Scholar]
- Lazaris, A.C.; Kavantzas, N.G.; Zorzos, H.S.; Tsavaris, N.V.; Davaris, P.S. Markers of drug resistance in relapsing colon cancer. J. Cancer Res. Clin. Oncol. 2002, 128, 114–118. [Google Scholar]
- Liang, P.; Pardee, A.B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 1992, 257, 967–971. [Google Scholar]
- Markova, V. Quantitative analysis of the expression of apoptosis-related genes. Folia Med. 1998, 40, 51–57. [Google Scholar]
- Yang, Z.; Woodahl, E.L.; Wang, X.Y.; Bui, T.; Shen, D.D.; Ho, R.J. Semi-quantitative RT-PCR method to estimate full-length mRNA levels of the multidrug resistance gene. Biotechniques 2002, 33. [Google Scholar]
- Gygi, S.P.; Rochon, Y.; Franza, B.R.; Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell Biol. 1999, 19, 1720–1730. [Google Scholar]
- MacBeath, G.; Schreiber, S.L. Printing proteins as microarrays for high-throughput function determination. Science 2000, 289, 1760–1763. [Google Scholar]
- Ge, H. A universal protein array system for quantitative detection of protein-protein, protein-DNA, protein-RNA and protein-ligand interactions. Nucleic Acids Res. 2000, 28, e3. [Google Scholar]
- Haab, B.B.; Dunham, M.J.; Brown, P.O. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001, 2, 1–13. [Google Scholar]
- Cras, J.J.; Rowe-Taitt, C.A.; Nivens, D.A.; Ligler, F.S. Comparison of chemical cleaning methods of glass in preparation for silanization. Bioelectrons 1999, 14, 683–688. [Google Scholar]
- Cole, S.R.; Aylett, G.W.; Harvey, N.L.; Cambareri, A.C.; Ashman, L.K. Increased expression of c-Kit or its ligand Steel Factor is not a common feature of adult acute myeloid leukaemia. Leukemia 1996, 10, 288–231. [Google Scholar]
- Piehler, J.; Brecht, A.; Hehl, K.; Gauglitz, G. Colloids and Surfaces. Biointerfaces 1999, 3, 325–336. [Google Scholar]
- Musto, P.; Melillo, L.; Lombardi, G. High risk of early resistant relapse for leukemia patients with presence of multidrug resistance associated P-glycoprotein positive cells in complete remission. Br. J. Haematol. 1991, 77, 50–53. [Google Scholar]
- Poeta, G.D.; Stasi, R.; Venditti, A. Prognostic value of cell marker analysis in denove acute myeloid leukemia. Leukemia 1994, 8, 38–41. [Google Scholar]
- Campos, L.; Guyotat, D.; Archimbaud, E. Clinical significance of multidrug resistance P-glycoprotein expression on acute non-lymphoblastic leukemia cells at diagnosis. Blood 1992, 79, 473–476. [Google Scholar]
- Ross, D.D.; Karp, J.E.; Chen, T.T. Expression of breast cancer resistance protein in blast cells from patients with acute leukemia. Blood 2000, 96, 365–368. [Google Scholar]


© 2005 by MDPI ( http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Du, J.; Chen, B.; Zhang, C.; Xu, X.; Cheng, J.; Gao, F.; Lu, Z. Protein Arrays for Multidrug-resistance in Human Leukemia Cell Determination. Sensors 2005, 5, 250-257. https://doi.org/10.3390/s5040259
Du J, Chen B, Zhang C, Xu X, Cheng J, Gao F, Lu Z. Protein Arrays for Multidrug-resistance in Human Leukemia Cell Determination. Sensors. 2005; 5(4):250-257. https://doi.org/10.3390/s5040259
Chicago/Turabian StyleDu, Juan, Baoan Chen, Chunxiu Zhang, Xiaoxing Xu, Jian Cheng, Feng Gao, and Zuhong Lu. 2005. "Protein Arrays for Multidrug-resistance in Human Leukemia Cell Determination" Sensors 5, no. 4: 250-257. https://doi.org/10.3390/s5040259



