An Investigation about Modern Deep Learning Strategies for Colon Carcinoma Grading
Abstract
1. Introduction
2. Related Work
3. Method and Data
3.1. Convolution Neural Networks (CNN)
3.1.1. ResNet
3.1.2. DenseNet
3.1.3. SENet
3.1.4. EfficientNet
3.1.5. RegNet
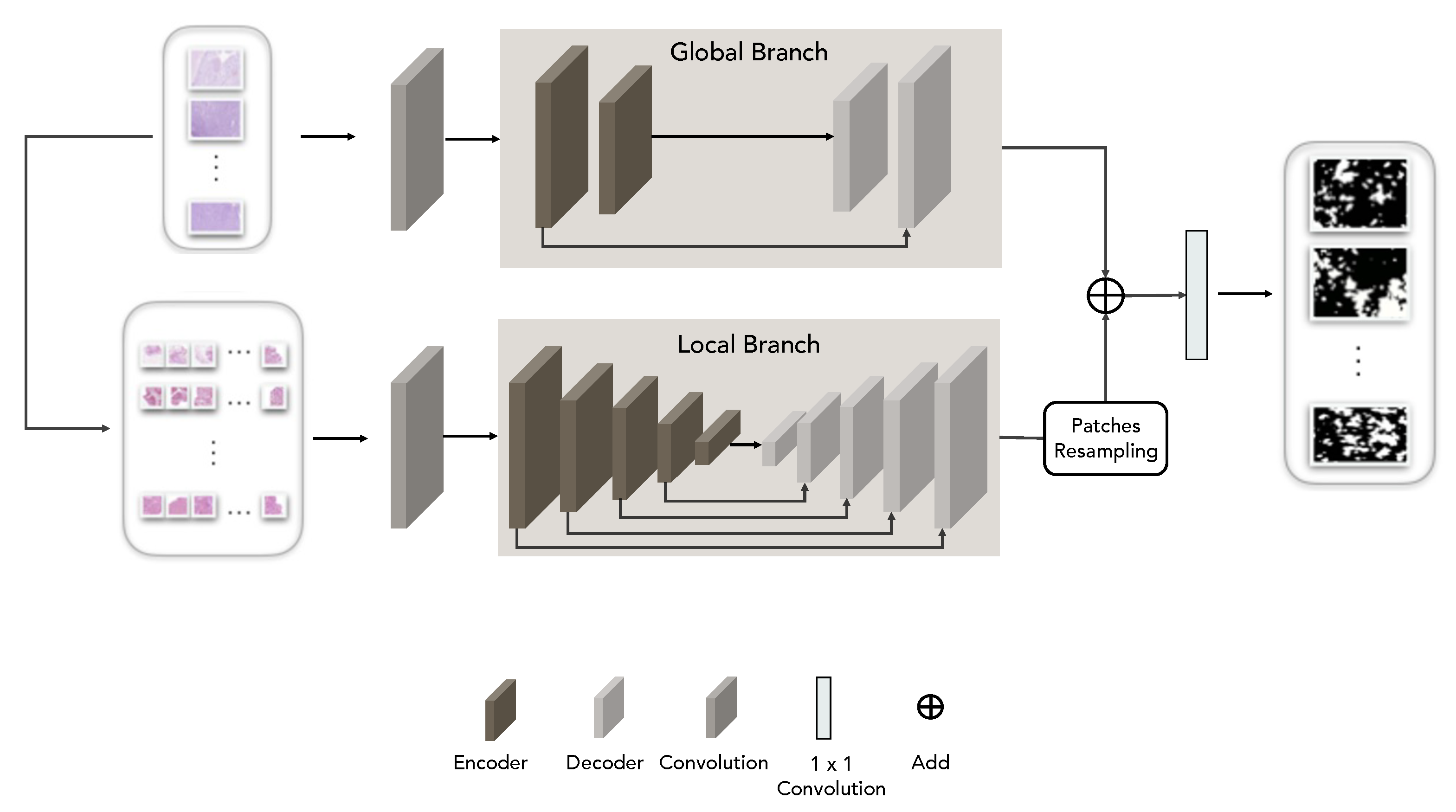
3.2. Transformer Networks
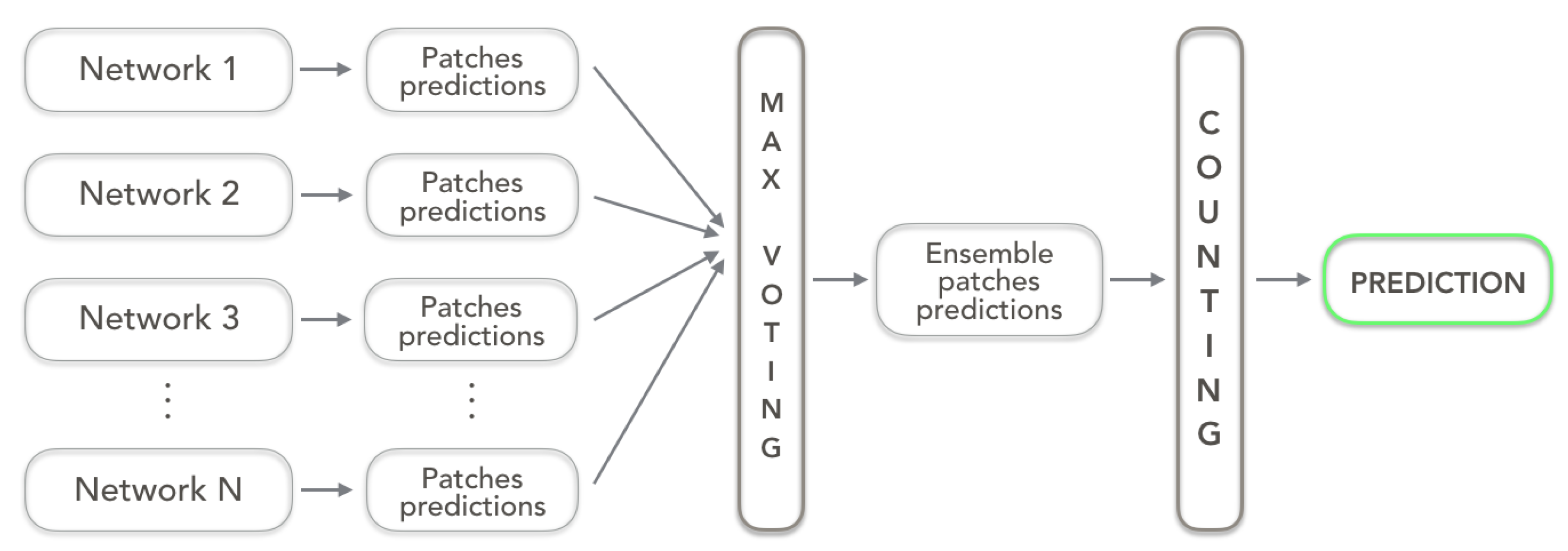
3.3. Ensemble Approach
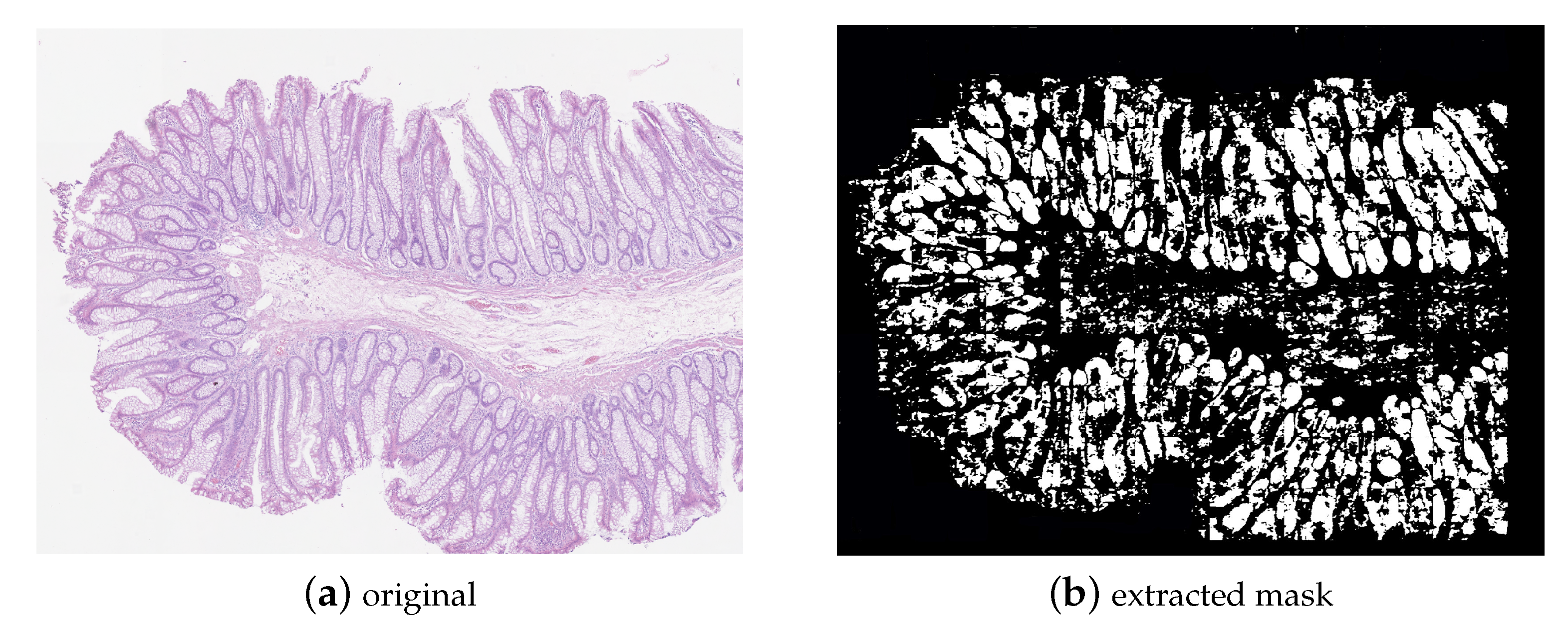
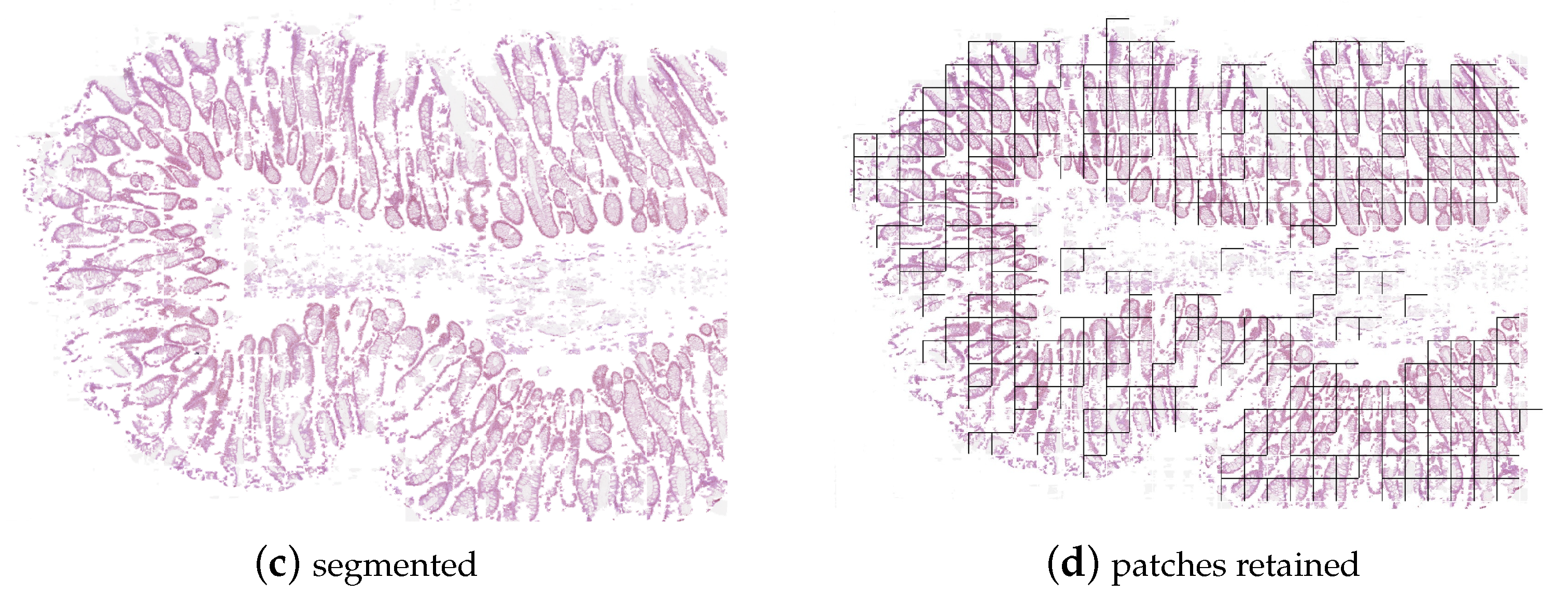
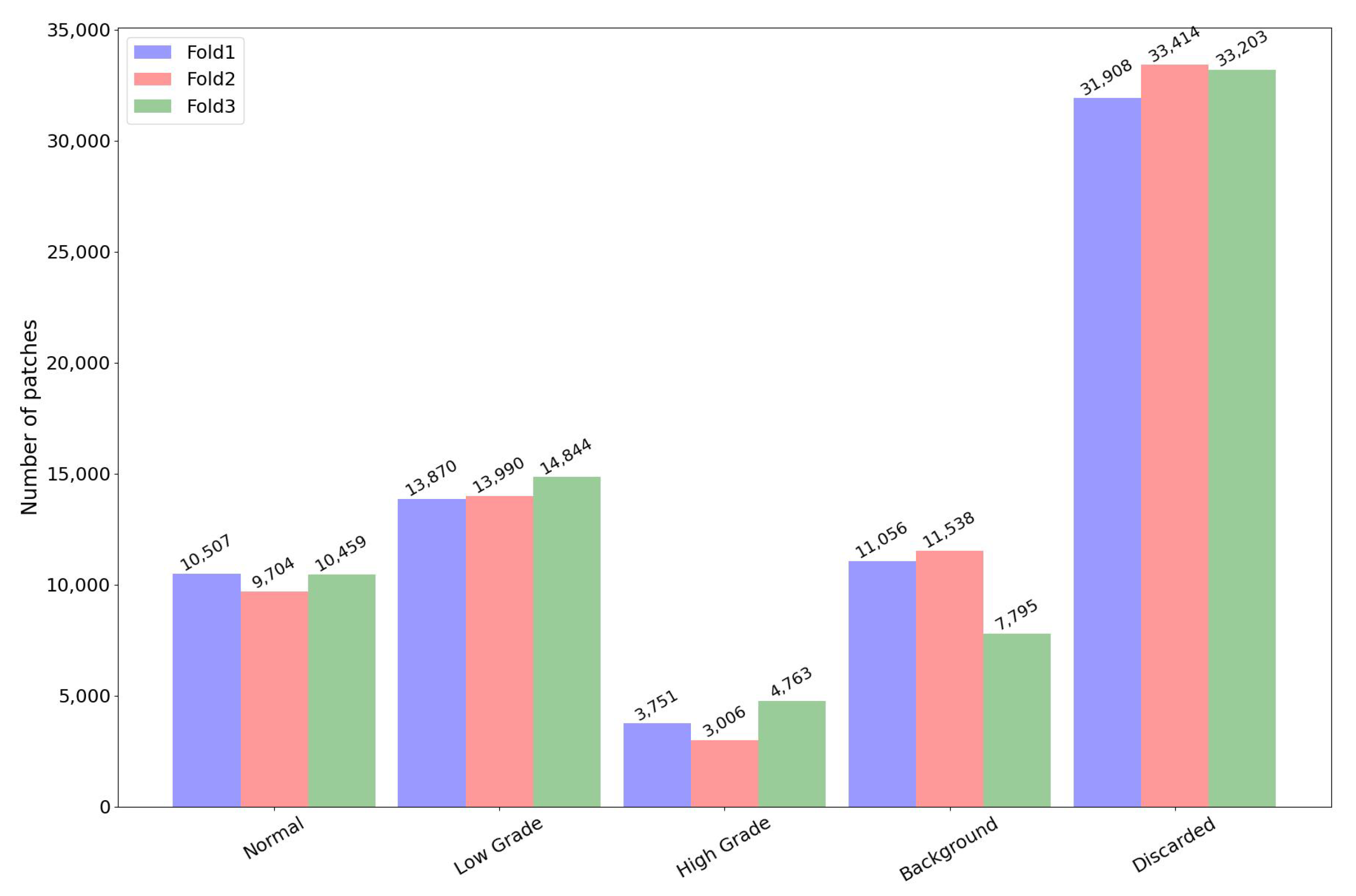
3.4. Datasets
4. Experimental Results
4.1. Transformer Model
4.2. Ensembles
5. Comparisons to Leading Approaches in the Literature
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Radosavovic, I.; Kosaraju, R.P.; Girshick, R.; He, K.; Dollár, P. Designing network design spaces. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10428–10436. [Google Scholar]
- Kong, B.; Li, Z.; Zhang, S. Toward large-scale histopathological image analysis via deep learning. In Biomedical Information Technology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 397–414. [Google Scholar]
- Saxena, S.; Gyanchandani, M. Machine learning methods for computer-aided breast cancer diagnosis using histopathology: A narrative review. J. Med. Imaging Radiat. Sci. 2020, 51, 182–193. [Google Scholar] [CrossRef]
- Das, A.; Nair, M.S.; Peter, S.D. Computer-aided histopathological image analysis techniques for automated nuclear atypia scoring of breast cancer: A review. J. Digit. Imaging 2020, 33, 1091–1121. [Google Scholar] [CrossRef] [PubMed]
- Carcagnì, P.; Leo, M.; Cuna, A.; Mazzeo, P.L.; Spagnolo, P.; Celeste, G.; Distante, C. Classification of skin lesions by combining multilevel learnings in a DenseNet architecture. In Image Analysis and Processing—ICIAP 2019, Proceedings of the International Conference on Image Analysis and Processing, Trento, Italy, 9–13 September 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 335–344. [Google Scholar]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Fleming, M.; Ravula, S.; Tatishchev, S.F.; Wang, H.L. Colorectal carcinoma: Pathologic aspects. J. Gastrointest. Oncol. 2012, 3, 153. [Google Scholar] [PubMed]
- Polikar, R. Ensemble learning. In Ensemble Machine Learning: Methods and Applications; Springer: Boston, MA, USA, 2012; pp. 1–34. [Google Scholar]
- Leo, M.; Farinella, G.M.; Furnari, A.; Medioni, G. Machine Vision for Assistive Technologies. Front. Comput. Sci. 2022, 70, 937433. [Google Scholar] [CrossRef]
- Müller, D.; Soto-Rey, I.; Kramer, F. An analysis on ensemble learning optimized medical image classification with deep convolutional neural networks. IEEE Access 2022, 10, 66467–66480. [Google Scholar] [CrossRef]
- Carcagnì, P.; Leo, M.; Celeste, G.; Distante, C.; Cuna, A. A systematic investigation on deep architectures for automatic skin lesions classification. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 8639–8646. [Google Scholar]
- Leo, M.; Farinella, G.M. Computer Vision for Assistive Healthcare; Academic Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 6000–6010. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J.; Shen, D. Transformers in medical image analysis: A review. Intell. Med. 2022, 3, 59–78. [Google Scholar] [CrossRef]
- Altunbay, D.; Cigir, C.; Sokmensuer, C.; Gunduz-Demir, C. Color graphs for automated cancer diagnosis and grading. IEEE Trans. Biomed. Eng. 2009, 57, 665–674. [Google Scholar] [CrossRef]
- Tosun, A.B.; Kandemir, M.; Sokmensuer, C.; Gunduz-Demir, C. Object-oriented texture analysis for the unsupervised segmentation of biopsy images for cancer detection. Pattern Recognit. 2009, 42, 1104–1112. [Google Scholar] [CrossRef]
- Awan, R.; Sirinukunwattana, K.; Epstein, D.; Jefferyes, S.; Qidwai, U.; Aftab, Z.; Mujeeb, I.; Snead, D.; Rajpoot, N. Glandular morphometrics for objective grading of colorectal adenocarcinoma histology images. Sci. Rep. 2017, 7, 16852. [Google Scholar] [CrossRef] [PubMed]
- Pacal, I.; Karaboga, D.; Basturk, A.; Akay, B.; Nalbantoglu, U. A comprehensive review of deep learning in colon cancer. Comput. Biol. Med. 2020, 126, 104003. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Samaras, D.; Kurc, T.M.; Gao, Y.; Davis, J.E.; Saltz, J.H. Patch-based convolutional neural network for whole slide tissue image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2424–2433. [Google Scholar]
- Vuong, T.L.T.; Lee, D.; Kwak, J.T.; Kim, K. Multi-task Deep Learning for Colon Cancer Grading. In Proceedings of the 2020 International Conference on Electronics, Information, and Communication (ICEIC), Barcelona, Spain, 19–22 January 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–2. [Google Scholar]
- Bychkov, D.; Linder, N.; Turkki, R.; Nordling, S.; Kovanen, P.E.; Verrill, C.; Walliander, M.; Lundin, M.; Haglund, C.; Lundin, J. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 2018, 8, 3395. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Alham, N.K.; Verrill, C.; Rittscher, J. Improving whole slide segmentation through visual context-a systematic study. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2018; pp. 192–200. [Google Scholar]
- Zhou, Y.; Graham, S.; Alemi Koohbanani, N.; Shaban, M.; Heng, P.; Rajpoot, N. CGC-Net: Cell Graph Convolutional Network for Grading of Colorectal Cancer Histology Images. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision Workshop (ICCVW), Seoul, Republic of Korea, 27–28 October 2019; pp. 388–398. [Google Scholar] [CrossRef]
- Shaban, M.; Awan, R.; Fraz, M.M.; Azam, A.; Tsang, Y.W.; Snead, D.; Rajpoot, N.M. Context-Aware Convolutional Neural Network for Grading of Colorectal Cancer Histology Images. IEEE Trans. Med. Imaging 2020, 39, 2395–2405. [Google Scholar] [CrossRef] [PubMed]
- Leo, M.; Carcagnì, P.; Signore, L.; Benincasa, G.; Laukkanen, M.O.; Distante, C. Improving Colon Carcinoma Grading by Advanced CNN Models. In Image Analysis and Processing—ICIAP 2022, Proceedings of the 21st International Conference, Lecce, Italy, 23–27 May 2022; Proceedings, Part I; Springer: Berlin/Heidelberg, Germany, 2022; pp. 233–244. [Google Scholar]
- Pei, Y.; Mu, L.; Fu, Y.; He, K.; Li, H.; Guo, S.; Liu, X.; Li, M.; Zhang, H.; Li, X. Colorectal tumor segmentation of CT scans based on a convolutional neural network with an attention mechanism. IEEE Access 2020, 8, 64131–64138. [Google Scholar] [CrossRef]
- Zhou, P.; Cao, Y.; Li, M.; Ma, Y.; Chen, C.; Gan, X.; Wu, J.; Lv, X.; Chen, C. HCCANet: Histopathological image grading of colorectal cancer using CNN based on multichannel fusion attention mechanism. Sci. Rep. 2022, 12, 15103. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Bengio, Y.; Simard, P.; Frasconi, P. Learning long-term dependencies with gradient descent is difficult. IEEE Trans. Neural Netw. 1994, 5, 157–166. [Google Scholar] [CrossRef]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 13–15 May 2010; JMLR Workshop and Conference Proceedings. pp. 249–256. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 448–456. [Google Scholar]
- Srivastava, R.K.; Greff, K.; Schmidhuber, J. Highway networks. arXiv 2015, arXiv:1505.00387. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Valanarasu, J.M.J.; Oza, P.; Hacihaliloglu, I.; Patel, V.M. Medical transformer: Gated axial-attention for medical image segmentation. arXiv 2021, arXiv:2102.10662. [Google Scholar]
- Ho, J.; Kalchbrenner, N.; Weissenborn, D.; Salimans, T. Axial attention in multidimensional transformers. arXiv 2019, arXiv:1912.12180. [Google Scholar]
- Awan, R.; Al-Maadeed, S.; Al-Saady, R.; Bouridane, A. Glandular structure-guided classification of microscopic colorectal images using deep learning. Comput. Electr. Eng. 2020, 85, 106450. [Google Scholar] [CrossRef]
- Shi, Q.; Katuwal, R.; Suganthan, P.; Tanveer, M. Random vector functional link neural network based ensemble deep learning. Pattern Recognit. 2021, 117, 107978. [Google Scholar] [CrossRef]
- Extended Colorectal Cancer Grading Dataset. Available online: https://warwick.ac.uk/fac/sci/dcs/research/tia/data/extended_crc_grading/ (accessed on 22 October 2020).
- Sirinukunwattana, K.; Pluim, J.P.; Chen, H.; Qi, X.; Heng, P.A.; Guo, Y.B.; Wang, L.Y.; Matuszewski, B.J.; Bruni, E.; Sanchez, U.; et al. Gland segmentation in colon histology images: The glas challenge contest. Med. Image Anal. 2017, 35, 489–502. [Google Scholar] [CrossRef]
- Kervadec, H.; Bouchtiba, J.; Desrosiers, C.; Granger, E.; Dolz, J.; Ayed, I.B. Boundary loss for highly unbalanced segmentation. Med. Image Anal. 2021, 67, 101851. [Google Scholar] [CrossRef]
- Chaddad, A.; Peng, J.; Xu, J.; Bouridane, A. Survey of Explainable AI Techniques in Healthcare. Sensors 2023, 23, 634. [Google Scholar] [CrossRef]



| Dataset | Normal | Low Grade | High Grade | Total |
|---|---|---|---|---|
| CRC | 71 | 33 | 35 | 139 |
| Extended-CRC | 120 | 120 | 60 | 300 |
| Model | Average (%) | Weighted (%) | Average (%) | Weighted (%) |
|---|---|---|---|---|
| (Binary) | (Binary) | (3-Classes) | (3-Classes) | |
| T+DenseNet121 | 98.33 ± 1.25 | 98.17 ± 1.30 | 88.91 ± 3.81 | 86.37 ± 4.12 |
| T+EfficientNet-B1 | 99.67 ± 0.47 | 99.72 ± 0.39 | 89.58 ± 4.17 | 87.50 ± 3.54 |
| T+EfficientNet-B2 | 98.66 ± 0.95 | 98.74 ± 0.91 | 89.92 ± 2.50 | 87.22 ± 2.08 |
| T+EfficientNet-B3 | 98.66 ± 0.95 | 98.89 ± 0.79 | 88.25 ± 2.55 | 84.44 ± 1.42 |
| T+EfficientNet-B4 | 97.65 ± 0.95 | 98.06 ± 0.79 | 88.92 ± 3.62 | 84.72 ± 3.36 |
| T+EfficientNet-B5 | 98.32 ± 1.72 | 98.46 ± 1.37 | 89.91 ± 5.42 | 88.05 ± 5.67 |
| T+EfficientNet-B7 | 97.66 ± 1.24 | 97.91 ± 0.89 | 88.59 ± 1.30 | 84.44 ± 0.38 |
| T+RegNetY6.4GF | 94.30 ± 2.07 | 94.57 ± 1.75 | 85.89 ± 3.64 | 81.94 ± 5.11 |
| T+RegNetY16GF | 97.66 ± 1.88 | 97.62 ± 1.56 | 88.57 ± 3.93 | 85.54 ± 3.76 |
| T+ResNet152 | 94.64 ± 2.04 | 94.82 ± 1.38 | 85.57 ± 1.31 | 81.36 ± 0.76 |
| T+SE-ResNet50 | 95.31 ± 1.24 | 95.52 ± 0.83 | 85.22 ± 3.37 | 79.70 ± 6.34 |
| Model | Average Training Time (min) | |
|---|---|---|
| Transformer | No Transformer | |
| DenseNet121 | 121.33 | 746.67 |
| EfficientNet-B1 | 121.33 | 452.67 |
| EfficientNet-B2 | 133.33 | 477.67 |
| EfficientNet-B3 | 168.00 | 480.67 |
| EfficientNet-B4 | 216.00 | 518.00 |
| EfficientNet-B5 | 309.00 | 677.67 |
| EfficientNet-B7 | 352.33 | 1188.00 |
| RegNetY6.4GF | 191.67 | 337.00 |
| RegNetY16GF | 349.00 | 699.33 |
| Resnet152 | 199.00 | 493.67 |
| SE-ResNet50 | 109.33 | 449.67 |
| Label | Models | Strategy |
|---|---|---|
| E1 | DenseNet121 EfficientNet-B7 RegNetY16GF | Max-Voting |
| E2 | DenseNet121 EfficientNet-B7 RegNetY16GF SE-ResNet50 | Max-Voting |
| E3 | DenseNet121 EfficientNet-B7 RegNetY16GF RegNetY6.4GF | Max-Voting |
| E4 | DenseNet121 EfficientNet-B7 RegNetY6.4GF | Max-Voting |
| E5 | DenseNet121 EfficientNet-B2 RegnetY16GF | Max-Voting |
| E6 | DeneNet121 EfficientNet-B2 RegNetY16GF | Max-Voting |
| E7 | DeneNet121 EfficientNet-B2 | Argmax |
| E8 | DenseNet121 EfficientNet-B7 RegNetY16GF SE-ResNet50 | Argmax |
| E9 | EfficientNet-B7 RegNetY16GF SE-ResNet50 | Argmax |
| E10 | DenseNet121 EfficientNet-B2 RegNetY16GF | Argmax |
| E11 | EfficientNet-B1 EfficientNet-B2 RegNetY16GF | Argmax |
| E12 | EfficientNet-B1 EfficientNet-B2 | Argmax |
| Ensemble | Average (%) | Weighted (%) | Average (%) | Weighted (%) |
|---|---|---|---|---|
| (Binary) | (Binary) | (3-Classes) | (3-Classes) | |
| E1 | 95.65 ± 1.87 | 95.52 ± 1.85 | 86.90 ± 4.16 | 84.15 ± 3.81 |
| E2 | 95.31 ± 2.48 | 95.68 ± 2.41 | 87.24 ± 3.37 | 83.88 ± 3.08 |
| E3 | 95.31 ± 1.68 | 95.40 ± 1.89 | 87.23 ± 4.18 | 84.15 ± 4.10 |
| E4 | 94.97 ± 1.62 | 95.12 ± 1.88 | 87.23 ± 1.18 | 84.15 ± 4.10 |
| E5 | 95.98 ± 2.45 | 95.81 ± 2.72 | 86.90 ± 4.39 | 84.15 ± 3.81 |
| E6 | 95.31 ± 2.34 | 95.40 ± 2.37 | 86.23 ± 3.37 | 83.32 ± 2.74 |
| E7 | 95.65 ± 2.05 | 95.82 ± 2.23 | 87.91 ± 3.33 | 84.72 ± 3.43 |
| E8 | 95.98 ± 2.15 | 95.95 ± 2.26 | 87.57 ± 3.75 | 84.71 ± 3.44 |
| E9 | 97.32 ± 1.26 | 97.33 ± 1.57 | 88.24 ± 4.26 | 85.53 ± 3.76 |
| T + E5 | 99.00 ± 0.82 | 99.02 ± 0.71 | 89.24 ± 4.09 | 87.49 ± 3.61 |
| T + E7 | 99.33 ± 0.94 | 99.44 ± 0.79 | 89.58 ± 3.83 | 87.22 ± 3.87 |
| T + E10 | 98.33 ± 1.25 | 98.46 ± 1.10 | 88.24 ± 4.10 | 85.52 ± 3.88 |
| T + E11 | 99.33 ± 0.94 | 99.44 ± 0.79 | 90.25 ± 3.74 | 88.06 ± 3.14 |
| T + E12 | 99.00 ± 0.82 | 99.02 ± 0.71 | 89.92 ± 3.00 | 87.49 ± 2.36 |
| Model | Average (%) | Weighted (%) | Average (%) | Weighted (%) |
|---|---|---|---|---|
| (Binary) | (Binary) | (3-Classes) | (3-Classes) | |
| Proposed | ||||
| T+EfficientNet-B1 | 99.67 ± 0.47 | 99.72 ± 0.39 | 89.58 ± 4.17 | 87.50 ± 3.54 |
| T+EfficientNet-B2 | 98.66 ± 0.95 | 98.74 ± 0.91 | 89.92 ± 2.50 | 87.22 ± 2.08 |
| T + E11 | 99.33 ± 0.94 | 99.44 ± 0.79 | 90.25 ± 3.74 | 88.06 ± 3.14 |
| Previous works | ||||
| ResNet50 [25] | ||||
| LR + LA-CNN [25] | ||||
| CNN-LSTM [23] | ||||
| CNN-SVM [20] | ||||
| CNN-LR [20] | ||||
| EfficientNet-B2 [26] | 96.99 ± 2.94 | 96.65 ± 3.11 | 87.58 ± 3.36 | 85.54 ± 2.21 |
| RegNetY-4.0GF [26] | 95.64 ± 0.94 | 95.37 ± 1.52 | 84.55 ± 2.57 | 81.36 ± 1.43 |
| RegNetY-6.4GF [26] | 94.31 ± 2.48 | 94.26 ± 2.15 | 86.57 ± 2.12 | 83.58 ± 2.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carcagnì, P.; Leo, M.; Signore, L.; Distante, C. An Investigation about Modern Deep Learning Strategies for Colon Carcinoma Grading. Sensors 2023, 23, 4556. https://doi.org/10.3390/s23094556
Carcagnì P, Leo M, Signore L, Distante C. An Investigation about Modern Deep Learning Strategies for Colon Carcinoma Grading. Sensors. 2023; 23(9):4556. https://doi.org/10.3390/s23094556
Chicago/Turabian StyleCarcagnì, Pierluigi, Marco Leo, Luca Signore, and Cosimo Distante. 2023. "An Investigation about Modern Deep Learning Strategies for Colon Carcinoma Grading" Sensors 23, no. 9: 4556. https://doi.org/10.3390/s23094556
APA StyleCarcagnì, P., Leo, M., Signore, L., & Distante, C. (2023). An Investigation about Modern Deep Learning Strategies for Colon Carcinoma Grading. Sensors, 23(9), 4556. https://doi.org/10.3390/s23094556










