Microfluidics and Nanomaterial-based Technologies for Circulating Tumor Cell Isolation and Detection
Abstract
1. Introduction
2. Circulating Tumor Cells (CTCs)
2.1. Formation of CTCs
2.2. Biological Properties of CTCs-Heterogeneity of CTCs
3. Application of Microfluidics Technology to CTC Isolation and Detection
3.1. Sorting Based on Size
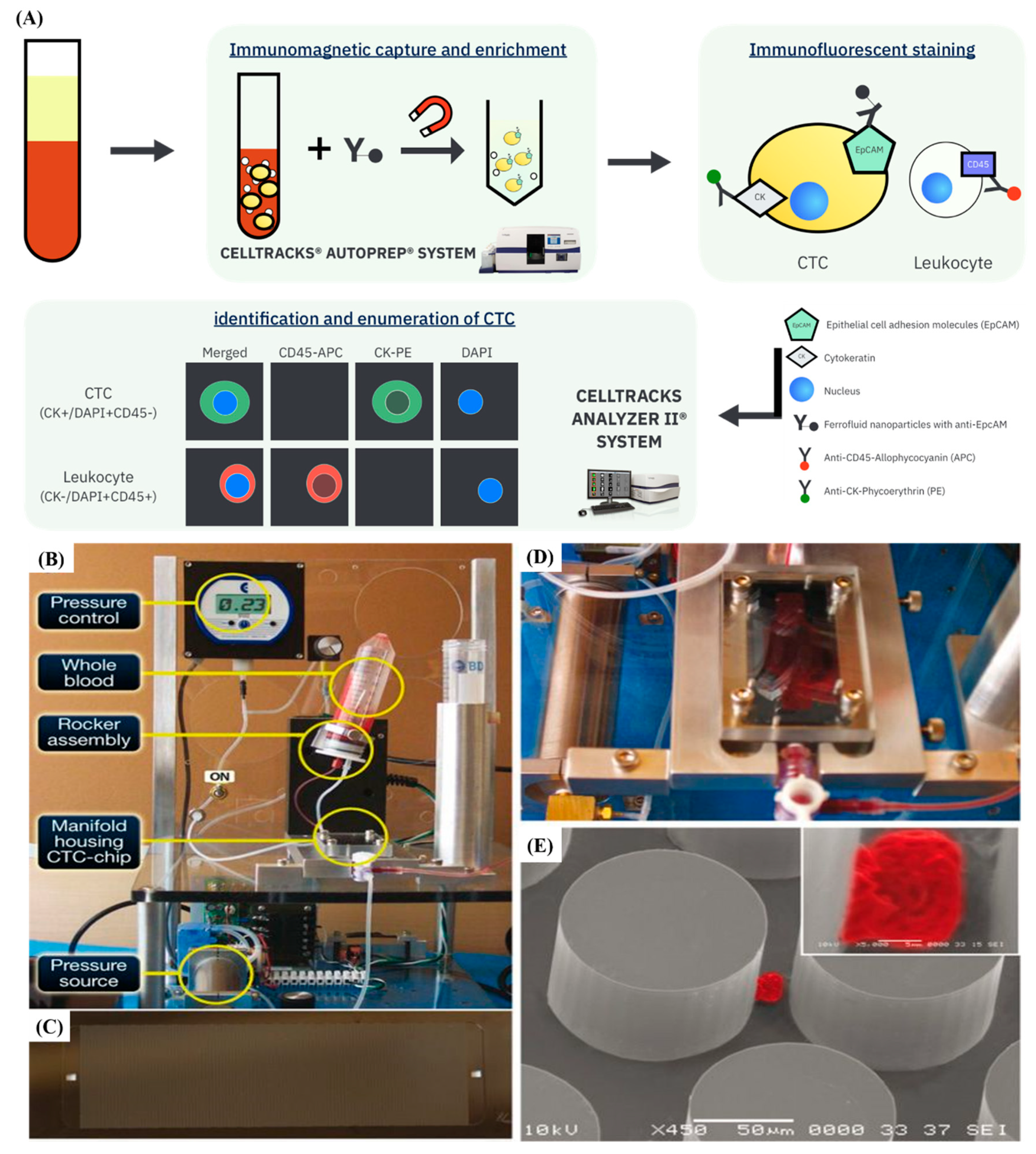
3.2. Immunoaffinity-Based CTCs Isolation and Detection
3.3. Dielectrophoresis (DEP)
4. Application of Nanomaterials in Microfluidic-Based Systems for Enhanced CTCs Isolation and Detection
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging biological principles of metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Cao, B.; Bray, F.; Beltrán-Sánchez, H.; Ginsburg, O.; Soneji, S.; Soerjomataram, I. Benchmarking life expectancy and cancer mortality: Global comparison with cardiovascular disease 1981–2010. BMJ 2017, 357, j2765. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zheng, R.; Xia, C.; Zeng, H.; Zhang, S.; Zou, X.; Yang, Z.; Li, H.; Chen, W. Interactions between life expectancy and the incidence and mortality rates of cancer in china: A population-based cluster analysis. Cancer Commun. 2018, 38, 44. [Google Scholar] [CrossRef]
- Marrugo-Ramírez, J.; Mir, M.; Samitier, J. Blood-based cancer biomarkers in liquid biopsy: A promising non-invasive alternative to tissue biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef]
- Smerage, J.B.; Barlow, W.E.; Hortobagyi, G.N.; Winer, E.P.; Leyland-Jones, B.; Srkalovic, G.; Tejwani, S.; Schott, A.F.; O’Rourke, M.A.; Lew, D.L.; et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: Swog s0500. J. Clin. Oncol. 2014, 32, 3483–3489. [Google Scholar] [CrossRef]
- Castro-Giner, F.; Gkountela, S.; Donato, C.; Alborelli, I.; Quagliata, L.; Ng, C.K.Y.; Piscuoglio, S.; Aceto, N. Cancer diagnosis using a liquid biopsy: Challenges and expectations. Diagnostics 2018, 8, 31. [Google Scholar] [CrossRef]
- Rack, B.K.; Schindlbeck, C.; Andergassen, U.; Schneeweiss, A.; Zwingers, T.; Lichtenegger, W.; Beckmann, M.; Sommer, H.L.; Pantel, K.; Janni, W.; et al. Use of circulating tumor cells (ctc) in peripheral blood of breast cancer patients before and after adjuvant chemotherapy to predict risk for relapse: The success trial. J. Clin. Oncol. 2010, 28, 1003. [Google Scholar] [CrossRef]
- Garrigós, N.; Gallego, J.; Guillén-Ponce, C.; Guaraz, P.; García-Bautista, M.; Castillejo, A.; Gómez-Martínez, Á.; Carrato, A.; Rodríguez-Lescure, Á.; Soto, J.L. Circulating tumour cell analysis as an early marker for relapse in stage ii and iii colorectal cancer patients: A pilot study. Clin. Transl. Oncol. 2010, 12, 142–147. [Google Scholar] [CrossRef]
- Ried, K.; Eng, P.; Sali, A. Screening for circulating tumour cells allows early detection of cancer and monitoring of treatment effectiveness: An observational study. Asian Pac. J. Cancer Prev. 2017, 18, 2275–2285. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Stopeck, A.; Terstappen, L.V.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with non-malignant diseases. J. Clin. Oncol. 2004, 22, 9552. [Google Scholar] [CrossRef][Green Version]
- Ashworth, T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust. Med. J. 1869, 14, 146–147. [Google Scholar]
- Chiang, S.P.H.; Cabrera, R.M.; Segall, J.E. Tumor cell intravasation. Am. J. Physiol. Cell Physiol. 2016, 311, C1–C14. [Google Scholar] [CrossRef]
- Gupta, G.P.; Massagué, J. Cancer metastasis: Building a framework. Cell 2006, 127, 679–695. [Google Scholar] [CrossRef]
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef]
- Andree, K.C.; van Dalum, G.; Terstappen, L.W.M.M. Challenges in circulating tumor cell detection by the cellsearch system. Mol. Oncol. 2016, 10, 395–407. [Google Scholar] [CrossRef]
- Li, Y.; Wu, S.; Bai, F. Molecular characterization of circulating tumor cells—From bench to bedside. Semin. Cell Dev. Biol. 2018, 75, 88–97. [Google Scholar] [CrossRef]
- Giancotti, F.G. Mechanisms governing metastatic dormancy and reactivation. Cell 2013, 155, 750–764. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial–mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef]
- Satelli, A.; Li, S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 2011, 68, 3033–3046. [Google Scholar] [CrossRef]
- Hazan, R.B.; Qiao, R.; Keren, R.; Badano, I.; Suyama, K. Cadherin switch in tumor progression. Ann. N. Y. Acad. Sci. 2004, 1014, 155–163. [Google Scholar] [CrossRef]
- Mego, M.; Mani, S.A.; Lee, B.N.; Li, C.; Evans, K.W.; Cohen, E.N.; Gao, H.; Jackson, S.A.; Giordano, A.; Hortobagyi, G.N.; et al. Expression of epithelial–mesenchymal transition-inducing transcription factors in primary breast cancer: The effect of neoadjuvant therapy. Int. J. Cancer 2012, 130, 808–816. [Google Scholar] [CrossRef]
- Diaz, M.T.d.M.; Abdallah, E.A.; Tariki, M.S.; Braun, A.C.; Dettino, A.L.A.; Nicolau, U.R.; Alves, V.d.S.; Chinen, L.T.D. Circulating tumor cells as marker of poor prognosis in metastatic lung cancer: A pilot study. Appl. Cancer Res. 2018, 38, 8. [Google Scholar] [CrossRef]
- Zhang, D.; Zhao, L.; Zhou, P.; Ma, H.; Huang, F.; Jin, M.; Dai, X.; Zheng, X.; Huang, S.; Zhang, T. Circulating tumor microemboli (ctm) and vimentin+ circulating tumor cells (ctcs) detected by a size-based platform predict worse prognosis in advanced colorectal cancer patients during chemotherapy. Cancer Cell Int. 2017, 17, 6. [Google Scholar] [CrossRef]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef]
- Au, S.H.; Storey, B.D.; Moore, J.C.; Tang, Q.; Chen, Y.L.; Javaid, S.; Sarioglu, A.F.; Sullivan, R.; Madden, M.W.; O’Keefe, R.; et al. Clusters of circulating tumor cells traverse capillary-sized vessels. Proc. Natl. Acad. Sci. USA 2016, 113, 4947–4952. [Google Scholar] [CrossRef]
- Duda, D.G.; Duyverman, A.M.; Kohno, M.; Snuderl, M.; Steller, E.J.; Fukumura, D.; Jain, R.K. Malignant cells facilitate lung metastasis by bringing their own soil. Proc. Natl. Acad. Sci. USA 2010, 107, 21677–21682. [Google Scholar] [CrossRef]
- Bithi, S.S.; Vanapalli, S.A. Microfluidic cell isolation technology for drug testing of single tumor cells and their clusters. Sci. Rep. 2017, 7, 41707. [Google Scholar] [CrossRef]
- Paoletti, C.; Miao, J.; Dolce, E.M.; Darga, E.P.; Repollet, M.I.; Doyle, G.V.; Gralow, J.R.; Hortobagyi, G.N.; Smerage, J.B.; Barlow, W.E.; et al. Circulating tumor cell clusters in metastatic breast cancer patients: A swog s0500 translational medicine study. Clin. Cancer Res. 2019, 25, 6089–6097. [Google Scholar] [CrossRef]
- Hou, J.M.; Krebs, M.G.; Lancashire, L.; Sloane, R.; Backen, A.; Swain, R.K.; Priest, L.J.C.; Greystoke, A.; Zhou, C.; Morris, K.; et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 2012, 30, 525–532. [Google Scholar] [CrossRef]
- Krebs, M.G.; Metcalf, R.L.; Carter, L.; Brady, G.; Blackhall, F.H.; Dive, C. Molecular analysis of circulating tumour cells—Biology and biomarkers. Nat. Rev. Clin. Oncol. 2014, 11, 129–144. [Google Scholar] [CrossRef]
- Coumans, F.A.W.; van Dalum, G.; Beck, M.; Terstappen, L.W.M.M. Filtration parameters influencing circulating tumor cell enrichment from whole blood. PLoS ONE 2013, 8, e61774. [Google Scholar] [CrossRef]
- Nedosekin, D.A.; Juratli, M.A.; Sarimollaoglu, M.; Moore, C.L.; Rusch, N.J.; Smeltzer, M.S.; Zharov, V.P.; Galanzha, E.I. Photoacoustic and photothermal detection of circulating tumor cells, bacteria and nanoparticles in cerebrospinal fluid in vivo and ex vivo. J. Biophotonics 2013, 6, 523–533. [Google Scholar] [CrossRef]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating tumor cells: Approaches to isolation and characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef]
- Barak, V.; Goike, H.; Panaretakis, K.W.; Einarsson, R. Clinical utility of cytokeratins as tumor markers. Clin. Biochem. 2004, 37, 529–540. [Google Scholar] [CrossRef]
- Onder, T.T.; Gupta, P.B.; Mani, S.A.; Yang, J.; Lander, E.S.; Weinberg, R.A. Loss of e-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008, 68, 3645–3654. [Google Scholar] [CrossRef]
- Runkle, E.A.; Mu, D. Tight junction proteins: From barrier to tumorigenesis. Cancer Lett. 2013, 337, 41–48. [Google Scholar] [CrossRef]
- Warzecha, C.C.; Shen, S.; Xing, Y.; Carstens, R.P. The epithelial splicing factors esrp1 and esrp2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 2009, 6, 546–562. [Google Scholar] [CrossRef]
- Ansieau, S.; Morel, A.P.; Hinkal, G.; Bastid, J.; Puisieux, A. Twisting an embryonic transcription factor into an oncoprotein. Oncogene 2010, 29, 3173–3184. [Google Scholar] [CrossRef]
- Barriere, G.; Riouallon, A.; Renaudie, J.; Tartary, M.; Rigaud, M. Mesenchymal and stemness circulating tumor cells in early breast cancer diagnosis. BMC Cancer 2012, 12, 114. [Google Scholar] [CrossRef]
- Hugo, H.J.; Pereira, L.; Suryadinata, R.; Drabsch, Y.; Gonda, T.J.; Gunasinghe, N.P.; Pinto, C.; Soo, E.T.; van Denderen, B.J.; Hill, P.; et al. Direct repression of myb by zeb1 suppresses proliferation and epithelial gene expression during epithelial-to-mesenchymal transition of breast cancer cells. Breast Cancer Res. 2013, 15, R113. [Google Scholar] [CrossRef]
- Sugimachi, K.; Yokobori, T.; Iinuma, H.; Ueda, M.; Ueo, H.; Shinden, Y.; Eguchi, H.; Sudo, T.; Suzuki, A.; Maehara, Y.; et al. Aberrant expression of plastin-3 via copy number gain induces the epithelial-mesenchymal transition in circulating colorectal cancer cells. Ann. Surg. Oncol. 2014, 21, 3680–3690. [Google Scholar] [CrossRef]
- Ribeiro-Samy, S.; Oliveira, M.I.; Pereira-Veiga, T.; Muinelo-Romay, L.; Carvalho, S.; Gaspar, J.; Freitas, P.P.; López-López, R.; Costa, C.; Diéguez, L. Fast and efficient microfluidic cell filter for isolation of circulating tumor cells from unprocessed whole blood of colorectal cancer patients. Sci. Rep. 2019, 9, 8032. [Google Scholar] [CrossRef]
- Vaidyanathan, R.; Soon, R.H.; Zhang, P.; Jiang, K.; Lim, C.T. Cancer diagnosis: From tumor to liquid biopsy and beyond. Lab Chip 2019, 19, 11–34. [Google Scholar] [CrossRef]
- Harouaka, R.A.; Nisic, M.; Zheng, S.-Y. Circulating tumor cell enrichment based on physical properties. J. Lab. Autom. 2013, 18, 455–468. [Google Scholar] [CrossRef]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef]
- Mohamed, H.; Murray, M.; Turner, J.N.; Caggana, M. Isolation of tumor cells using size and deformation. J. Chromatogr. A 2009, 1216, 8289–8295. [Google Scholar] [CrossRef]
- Zheng, S.; Lin, H.K.; Lu, B.; Williams, A.; Datar, R.; Cote, R.J.; Tai, Y.-C. 3d microfilter device for viable circulating tumor cell (ctc) enrichment from blood. Biomed. Microdevices 2011, 13, 203–213. [Google Scholar] [CrossRef]
- Hur, S.C.; Mach, A.J.; Di Carlo, D. High-throughput size-based rare cell enrichment using microscale vortices. Biomicrofluidics 2011, 5, 22206. [Google Scholar] [CrossRef]
- Renier, C.; Pao, E.; Che, J.; Liu, H.E.; Lemaire, C.A.; Matsumoto, M.; Triboulet, M.; Srivinas, S.; Jeffrey, S.S.; Rettig, M.; et al. Label-free isolation of prostate circulating tumor cells using vortex microfluidic technology. NPJ Precis. Oncol. 2017, 1, 15. [Google Scholar] [CrossRef]
- Huang, L.R.; Cox, E.C.; Austin, R.H.; Sturm, J.C. Continuous particle separation through deterministic lateral displacement. Science 2004, 304, 987–990. [Google Scholar] [CrossRef]
- Salafi, T.; Zhang, Y.; Zhang, Y. A review on deterministic lateral displacement for particle separation and detection. Nano-Micro. Lett. 2019, 11, 77. [Google Scholar] [CrossRef]
- Okano, H.; Konishi, T.; Suzuki, T.; Suzuki, T.; Ariyasu, S.; Aoki, S.; Abe, R.; Hayase, M. Enrichment of circulating tumor cells in tumor-bearing mouse blood by a deterministic lateral displacement microfluidic device. Biomed. Microdevices 2015, 17, 9964. [Google Scholar] [CrossRef]
- Fachin, F.; Spuhler, P.; Martel-Foley, J.M.; Edd, J.F.; Barber, T.A.; Walsh, J.; Karabacak, M.; Pai, V.; Yu, M.; Smith, K.; et al. Monolithic chip for high-throughput blood cell depletion to sort rare circulating tumor cells. Sci. Rep. 2017, 7, 10936. [Google Scholar] [CrossRef]
- Au, S.H.; Edd, J.; Stoddard, A.E.; Wong, K.H.K.; Fachin, F.; Maheswaran, S.; Haber, D.A.; Stott, S.L.; Kapur, R.; Toner, M. Microfluidic isolation of circulating tumor cell clusters by size and asymmetry. Sci. Rep. 2017, 7, 2433. [Google Scholar] [CrossRef]
- Shen, Z.; Wu, A.; Chen, X. Current detection technologies for circulating tumor cells. Chem. Soc. Rev. 2017, 46, 2038–2056. [Google Scholar] [CrossRef]
- Kulasinghe, A.; Wu, H.; Punyadeera, C.; Warkiani, M.E. The use of microfluidic technology for cancer applications and liquid biopsy. Micromachines 2018, 9, 397. [Google Scholar] [CrossRef]
- Lowes, L.E.; Hedley, B.D.; Keeney, M.; Allan, A.L. User-defined protein marker assay development for characterization of circulating tumor cells using the cellsearch® system. Cytom. A 2012, 81A, 983–995. [Google Scholar] [CrossRef]
- Tibbe, A.G.J.; Miller, M.C.; Terstappen, L.W.M.M. Statistical considerations for enumeration of circulating tumor cells. Cytom. A 2007, 71A, 154–162. [Google Scholar] [CrossRef]
- Estes, M.D.; Ouyang, B.; Ho, S.-m.; Ahn, C.H. Isolation of prostate cancer cell subpopulations of functional interest by use of an on-chip magnetic bead-based cell separator. J. Micromech. Microeng. 2009, 19, 095015. [Google Scholar] [CrossRef]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef]
- Myung, J.H.; Eblan, M.J.; Caster, J.M.; Park, S.J.; Poellmann, M.J.; Wang, K.; Tam, K.A.; Miller, S.M.; Shen, C.; Chen, R.C.; et al. Multivalent binding and biomimetic cell rolling improves the sensitivity and specificity of circulating tumor cell capture. Clin. Cancer Res. 2018, 24, 2539–2547. [Google Scholar] [CrossRef]
- Stott, S.L.; Hsu, C.-H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kim, T.H.; Zhang, Z.; Azizi, E.; Pham, T.M.; Paoletti, C.; Lin, J.; Ramnath, N.; Wicha, M.S.; Hayes, D.F.; et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 2013, 8, 735–741. [Google Scholar] [CrossRef]
- Voldman, J. Electrical forces for microscale cell manipulation. Annu. Rev. Biomed. Eng. 2006, 8, 425–454. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Shim, S. Isolation of circulating tumor cells by dielectrophoresis. Cancers 2014, 6, 545–579. [Google Scholar] [CrossRef]
- Alshareef, M.; Metrakos, N.; Juarez Perez, E.; Azer, F.; Yang, F.; Yang, X.; Wang, G. Separation of tumor cells with dielectrophoresis-based microfluidic chip. Biomicrofluidics 2013, 7, 11803. [Google Scholar] [CrossRef]
- Becker, F.F.; Wang, X.B.; Huang, Y.; Pethig, R.; Vykoukal, J.; Gascoyne, P.R. Separation of human breast cancer cells from blood by differential dielectric affinity. Proc. Natl. Acad. Sci. USA 1995, 92, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Aghaamoo, M.; Aghilinejad, A.; Chen, X.; Xu, J. On the design of deterministic dielectrophoresis for continuous separation of circulating tumor cells from peripheral blood cells. Electrophoresis 2019, 40, 1486–1493. [Google Scholar] [CrossRef]
- Biju, V. Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem. Soc. Rev. 2014, 43, 744–764. [Google Scholar] [CrossRef]
- Rolfe, P. Micro- and nanosensors for medical and biological measurement. Sens. Mater. 2012, 24, 275–302. [Google Scholar] [CrossRef]
- Von Maltzahn, G.; Park, J.-H.; Lin, K.Y.; Singh, N.; Schwöppe, C.; Mesters, R.; Berdel, W.E.; Ruoslahti, E.; Sailor, M.J.; Bhatia, S.N. Nanoparticles that communicate in vivo to amplify tumour targeting. Nat. Mater. 2011, 10, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chen, J.; Li, X.; Zhao, Y.; Zughaier, S.M. Culture-free diagnostics of pseudomonas aeruginosa infection by silver nanorod array based sers from clinical sputum samples. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 1863–1870. [Google Scholar] [CrossRef]
- Dreyer, D.R.; Park, S.; Bielawski, C.W.; Ruoff, R.S. The chemistry of graphene oxide. Chem. Soc. Rev. 2010, 39, 228–240. [Google Scholar] [CrossRef]
- Cheng, S.J.; Chiu, H.Y.; Kumar, P.V.; Hsieh, K.Y.; Yang, J.W.; Lin, Y.R.; Shen, Y.C.; Chen, G.Y. Simultaneous drug delivery and cellular imaging using graphene oxide. Biomater. Sci. 2018, 6, 813–819. [Google Scholar] [CrossRef]
- Yang, J.W.; Hsieh, K.Y.; Kumar, P.V.; Cheng, S.J.; Lin, Y.R.; Shen, Y.C.; Chen, G.Y. Enhanced osteogenic differentiation of stem cells on phase-engineered graphene oxide. ACS Appl. Mater. Interfaces 2018, 10, 12497–12503. [Google Scholar] [CrossRef]
- Yang, J.W.; Tseng, M.L.; Fu, Y.M.; Kang, C.H.; Cheng, Y.T.; Kuo, P.H.; Tzeng, C.K.; Chiou, S.H.; Wu, C.Y.; Chen, G.Y. Printable graphene oxide micropatterns for a bio-subretinal chip. Adv. Healthc. Mater. 2018, 7, 1800365. [Google Scholar] [CrossRef] [PubMed]
- Pramani, K.A.; Jones, S.; Gao, Y.; Sweet, C.; Vangara, A.; Begum, S.; Ray, P.C. Multifunctional hybrid graphene oxide for circulating tumor cell isolation and analysis. Adv. Drug Deliv. Rev. 2018, 125, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Yoon, H.J.; Fouladdel, S.; Wang, Y.; Kozminsky, M.; Burness, M.L.; Paoletti, C.; Zhao, L.; Azizi, E.; Wicha, M.S.; et al. Characterizing circulating tumor cells isolated from metastatic breast cancer patients using graphene oxide based microfluidic assay. Adv. Biosyst. 2019, 3, 1800278. [Google Scholar] [CrossRef]
- Wu, Y.; Xue, P.; Kang, Y.; Hui, K.M. Highly specific and ultrasensitive graphene-enhanced electrochemical detection of low-abundance tumor cells using silica nanoparticles coated with antibody-conjugated quantum dots. Anal. Chem. 2013, 85, 3166–3173. [Google Scholar] [CrossRef]
- Sonawane, M.D.; Nimse, S.B. Surface modification chemistries of materials used in diagnostic platforms with biomolecules. J. Chem. 2016, 2016, 9241378. [Google Scholar] [CrossRef]
- O’connell, M.J. Carbon Nanotubes: Properties and Applications; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Marshall, M.W.; Popa-Nita, S.; Shapter, J.G. Measurement of functionalised carbon nanotube carboxylic acid groups using a simple chemical process. Carbon 2006, 44, 1137–1141. [Google Scholar] [CrossRef]
- Tam, P.D.; Van Hieu, N.; Chien, N.D.; Le, A.-T.; Anh Tuan, M. DNA sensor development based on multi-wall carbon nanotubes for label-free influenza virus (type a) detection. J. Immunol. Methods 2009, 350, 118–124. [Google Scholar] [CrossRef]
- Eitan, A.; Jiang, K.; Dukes, D.; Andrews, R.; Schadler, L.S. Surface modification of multiwalled carbon nanotubes: Toward the tailoring of the interface in polymer composites. Chem. Mater. 2003, 15, 3198–3201. [Google Scholar] [CrossRef]
- Dontha, N.; Nowall, W.B.; Kuhr, W.G. Generation of biotin/avidin/enzyme nanostructures with maskless photolithography. Anal. Chem. 1997, 69, 2619–2625. [Google Scholar] [CrossRef]
- Wilchek, M.; Bayer, E.A. The avidin-biotin complex in bioanalytical applications. Anal. Biochem. 1988, 171, 1–32. [Google Scholar] [CrossRef]
- Reimhult, E.; Höök, F. Design of surface modifications for nanoscale sensor applications. Sensors 2015, 15, 1635–1675. [Google Scholar] [CrossRef] [PubMed]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Nwe, K.; Brechbiel, M.W. Growing applications of “click chemistry” for bioconjugation in contemporary biomedical research. Cancer Biother. Radiopharm. 2009, 24, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise huisgen cycloaddition process: Copper(i)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Huisgen, R. 1,3-dipolar cycloadditions. Past and future. Angew. Chem. Int. Ed. Engl. 1963, 2, 565–598. [Google Scholar] [CrossRef]
- Wang, Q.; Chan, T.R.; Hilgraf, R.; Fokin, V.V.; Sharpless, K.B.; Finn, M.G. Bioconjugation by copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J. Am. Chem. Soc. 2003, 125, 3192–3193. [Google Scholar] [CrossRef]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A strain-promoted [3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef]
- Eeftens, J.M.; van der Torre, J.; Burnham, D.R.; Dekker, C. Copper-free click chemistry for attachment of biomolecules in magnetic tweezers. BMC Biophys. 2015, 8, 9. [Google Scholar] [CrossRef]
- Bardhan, N.M.; Kumar, P.V.; Li, Z.; Ploegh, H.L.; Grossman, J.C.; Belcher, A.M.; Chen, G.-Y. Enhanced cell capture on functionalized graphene oxide nanosheets through oxygen clustering. ACS Nano 2017, 11, 1548–1558. [Google Scholar] [CrossRef]
- Trilling, A.K.; Hesselink, T.; Houwelingen, A.v.; Cordewener, J.H.G.; Jongsma, M.A.; Schoffelen, S.; Hest, J.C.M.v.; Zuilhof, H.; Beekwilder, J. Orientation of llama antibodies strongly increases sensitivity of biosensors. Biosens. Bioelectron. 2014, 60, 130–136. [Google Scholar] [CrossRef]
- Trilling, A.K.; Harmsen, M.M.; Ruigrok, V.J.B.; Zuilhof, H.; Beekwilder, J. The effect of uniform capture molecule orientation on biosensor sensitivity: Dependence on analyte properties. Biosens. Bioelectron. 2013, 40, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, N.; Liu, H.; Chen, C.; Ding, P.; Yue, X.; Zou, H.; Xing, C.; Pei, R. High-efficiency isolation and rapid identification of heterogeneous circulating tumor cells (ctcs) using dual-antibody-modified fluorescent-magnetic nanoparticles. ACS Appl. Mater. Interfaces 2019, 11, 39586–39593. [Google Scholar] [CrossRef] [PubMed]






| Phenotype | Marker | Reference |
|---|---|---|
| Epithelial-like | EpCAM | [37] |
| Cytokeratin | [38] | |
| E-cadherin | [39] | |
| Zonula occludens | [40] | |
| Epithelial splicing regulator1 | [41] | |
| Mesenchymal-like | Vimentin | [23] |
| N-cadherin | [24] | |
| Twist1 | [42,43] | |
| ZEB1 | [44] | |
| Plastin-3 | [45] |
| Strategies | Technology | Selection Criteria | Biomarkers used for CTCs Characterization after Processing | Throughput | Performance in Spiking Experiment | Performance in Clinical Samples | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| Sample | Capture Efficiency (Captured CTCs/Total CTCs in Samples) | Sample | Sensitivity (Captured CTCs/Total Volume of Sample or Total) | ||||||
| Microfluidics - DLD | monolithic CTC-iChip | Size cutoff of 3.8 µm and CD45-/CD16-/CD66- | DRAQ5+/EpCAM+ (Prostate-Lung-Breast)/ CK/Her2 (Breast)/ CD146/NG2+ (Melanoma) | ~15 million cells/second | 11 different cell lines (SkMel28, H1650, H1975, H3122, LNCAP, PC3, PC3-9, VCAP, MB231, MCF-7, SkBR), spiked in 1X PBS with 1% F68 (average concentration: 425 cells/mL) | 99.5% | Whole blood from cancer patients | 1~63 CTCs/ mL | [57] |
| cancer-specific antibodies-coated magnetic beads | CellSearch | EpCAM+ | CD45-/CK+/DAPI+ | - | 1000 or 4000 human tumor cells spiked in 7.5 mL whole blood, diluted with 6.5 mL of dilution buffer (Veridex) | - | - | - | [61] |
| cancer-specific antibodies-coated magnetic beads | On-chip magnetic bead-based cell separator | PSMA+/CD10− | - | 2-200 µL/ min | 1 million cells in 100 µL PBS buffer | 60 ± 10% | - | - | [63] |
| Microfluidics + cancer-specific antibodies-coated microposts | CTC-chip | EpCAM+ | CK+/CD45-/DAPI+ | 1~2 mL/ h | Whole blood spiked with cancer cells (concentration ranging from 50 to 50,000 tumor cells per mL) | >60% | Whole blood from cancer patients | 1 target cell/ 2.7 mL | [64] |
| cancer-specific antibodies immobilized-surfaces functionalized with dendrimers | CapioCyte | E-selectin-/EpCAM+/HER-2+/EGFR+ | CK+/ CD45-/ DAPI+ | 25 µL/ min | - | - | Whole blood from cancer patients | 19~849 CTCs/ mL | [65] |
| Cancer-specific antibodies-coated microposts | HB Chip | EpCAM+ | CK+/CD45-/PSA+/CEPX+/AR+/DAPI+ | 1.5~2.5 mL/h | PC3 cells spiked into whole blood | 91.8% ± 5.2% | Whole blood from metastatic prostate cancer patients at various stages of treatment | 12~3167 CTCs/mL | [66] |
| Nanomaterial-interfaced patterned gold surface | GO-interfaced patterned gold surface | EpCAM+ | CK+/CD45- | 1~3 mL/ h | Fluorescent tracker dyed MCF-7 and PC-3 spiked into buffer | 82.3% | Whole blood samples from patients with metastatic breast cancer, early stage lung cancer and metastatic pancreatic cancer | 1~23 CTCs/ mL | [67] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, S.-J.; Hsieh, K.Y.; Chen, S.-L.; Chen, C.-Y.; Huang, C.-Y.; Tsou, H.-I.; Kumar, P.V.; Hsieh, J.C.-H.; Chen, G.-Y. Microfluidics and Nanomaterial-based Technologies for Circulating Tumor Cell Isolation and Detection. Sensors 2020, 20, 1875. https://doi.org/10.3390/s20071875
Cheng S-J, Hsieh KY, Chen S-L, Chen C-Y, Huang C-Y, Tsou H-I, Kumar PV, Hsieh JC-H, Chen G-Y. Microfluidics and Nanomaterial-based Technologies for Circulating Tumor Cell Isolation and Detection. Sensors. 2020; 20(7):1875. https://doi.org/10.3390/s20071875
Chicago/Turabian StyleCheng, Sheng-Jen, Kuan Yu Hsieh, Shiue-Luen Chen, Chong-You Chen, Chien-Yu Huang, Hung-I Tsou, Priyank V. Kumar, Jason Chia-Hsun Hsieh, and Guan-Yu Chen. 2020. "Microfluidics and Nanomaterial-based Technologies for Circulating Tumor Cell Isolation and Detection" Sensors 20, no. 7: 1875. https://doi.org/10.3390/s20071875
APA StyleCheng, S.-J., Hsieh, K. Y., Chen, S.-L., Chen, C.-Y., Huang, C.-Y., Tsou, H.-I., Kumar, P. V., Hsieh, J. C.-H., & Chen, G.-Y. (2020). Microfluidics and Nanomaterial-based Technologies for Circulating Tumor Cell Isolation and Detection. Sensors, 20(7), 1875. https://doi.org/10.3390/s20071875





