A Near-infrared Turn-on Fluorescent Sensor for Sensitive and Specific Detection of Albumin from Urine Samples
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthetic Procedure of DMAT-π-CAP (5)
2.1.1. Synthesis of 5-(dimethylamino) thiophene-2-carbaldehyde (2)
2.1.2. Synthesis of (E)-3-(5-(dimethylamino) thiophen-2-yl) acrylaldehyde (3)
2.1.3. Synthesis of (2E, 4E)-5-(5-(dimethylamino)thiophen-2-yl)penta-2,4-dienal (4)
2.1.4. Synthesis of (2E,4E,6E)-2-(3-chlorobenzoyl)-7-(5-(dimethylamino)thiophen-2-yl)hepta-2,4,6- trienenitrile (5)
2.2. Determination of Fluorescence Properties of DMAT-π-CAP in PBS Buffer (pH 7.4)
2.3. Determination of the Quantum Efficiency of Fluorescence of DMAT-π-CAP
2.4. HSA-Selective Turn-on Fluorescence of DMAT-π-CAP
2.5. Time-Dependent Fluorescence of DMAT-π-CAP after Incubation with HSA.
2.6. Concentration-Dependent Fluorescence of DMAT-π-CAP after Incubation with HSA
2.7. Limit of Detection (LOD) of DMAT-π-CAP
2.8. Job’s Plot Analysis of DMAT-π-CAP
2.9. Dissociation Constant (Kd) of DMAT-π-CAP from HSA
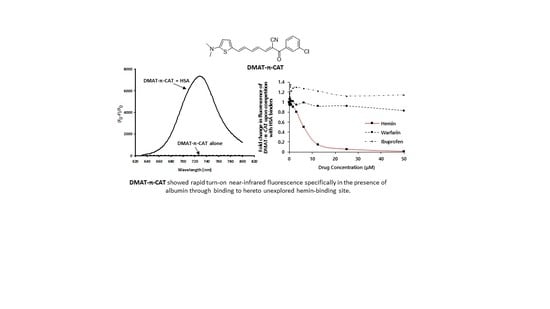
2.10. Assignment of the Binding Site of DMAT-π-CAP on HSA
2.11. Assessment of Urinary Albumin Levels
2.11.1. Fluorometric Analysis of Urinary Albumin with DMAT-π-CAP
2.11.2. Determination of Urinary Albumin by Immunoassay
2.12. Statistics
3. Results
3.1. Synthesis of DMAT-π-CAP
3.2. Near-infrared Turn-on Fluorescence Properties of DMAT-π-CAP
3.3. The Binding Properties of DMAT-π-CAP to HSA
3.4. Determination of Urinary HSA Levels by Using DMAT-π-CAP
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Quinlan, G.J.; Martin, G.S.; Evans, T.W. Albumin: Biochemical properties and therapeutic potential. Hepatology 2005, 41, 1211–1219. [Google Scholar] [CrossRef]
- Mazzaferro, E.M.; Rudloff, E.; Kirby, R. The role of albumin replacement in the critically ill veterinary patient. J. Vet. Emerg. Crit. Care 2002, 12, 113–124. [Google Scholar] [CrossRef]
- Arques, S.; Ambrosi, P.J. Human serum albumin in the clinical syndrome of heart failure. Card. Fail. 2011, 17, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.; Katz, R.; Ix, J.H.; Gutierrez, O.M.; Peralta, C.A.; Parikh, C.R.; Satterfield, S.; Petrovic, S.; Devarajan, P.; Bennett, M.; et al. Association of serum albumin levels with kidney function decline and incident chronic kidney disease in elders. Nephrol. Dial. Transplant 2018, 33, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Levitt, D.G.; Levitt, M.D. Protein losing enteropathy: comprehensive review of the mechanistic association with clinical and subclinical disease states. Clin. Exp. Gastroenterol. 2017, 10, 147–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doweiko, J.P.; Nompleggi, D.J. The role of albumin in human physiology and pathophysiology, Part III: Albumin and disease states. J. Parenter. Enteral Nutr. 1991, 15, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Levitt, D.G.; Levitt, M.D. Human serum albumin homeostasis: A new look at the roles of synthesis, catabolism, renal and gastrointestinal excretion, and the clinical value of serum albumin measurements. Int. J. Gen. Med. 2016, 9, 229–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sand, K.M.; Bern, M.; Nilsen, J.; Noordzij, H.T.; Sandlie, I.; Andersen, J.T. Unraveling the interaction between FcRn and albumin: Opportunities for design of albumin-based therapeutics. Front. Immunol. 2015, 5, 682. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, K.; Chuang, V.T.; Maruyama, T.; Otagiri, M. Albumin–drug interaction and its clinical implication. Biochim. Biophys. Acta 2013, 1830, 5435–5443. [Google Scholar] [CrossRef]
- Dennis, M.S.; Zhang, M.; Meng, Y.G.; Kadkhodayan, M.; Kirchhofer, D.; Combs, D.; Damico, L.A. Albumin binding as a general strategy for improving the pharmacokinetics of proteins. J. Biol. Chem. 2002, 277, 35035–35043. [Google Scholar] [CrossRef] [Green Version]
- Seegmiller, J.C.; Sviridov, D.; Larson, T.S.; Borland, T.M.; Hortin, G.L.; Lieske, J.C. Comparison of urinary albumin quantification by immunoturbidimetry, competitive immunoassay, and protein-cleavage liquid chromatography-tandem mass spectrometry. Clin. Chem. 2009, 55, 1991–1994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasanayagam, L.J.; Lim, K.L.; Beng, C.G.; Lau, K.S. Measurement of urine albumin using bromocresol green. Clin. Chim. Acta 1973, 44, 53–57. [Google Scholar] [CrossRef]
- Choi, S.; Choi, E.Y.; Kim, D.J.; Kim, J.H.; Kim, T.S.; Oh, S.W. A rapid, simple measurement of human albumin in whole blood using a fluorescence immunoassay (I). Clin. Chim. Acta 2004, 339, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Rajasekhar, K.; Achar, C.J.; Govindaraju, T. A red-NIR emissive probe for the selective detection of albumin in urine samples and live cells. Org. Biomol. Chem. 2017, 15, 1584–1588. [Google Scholar] [CrossRef]
- Wang, Y.-R.; Feng, L.; Xu, L.; Li, Y.; Wang, D.- D.; Hou, J.; Zhou, K.; Jin, Q.; Ge, G.-B.; Cui, J.-N.; et al. A rapid-response fluorescent probe for the sensitive and selective detection of human albumin in plasma and cell culture supernatants. Chem. Commun. 2016, 52, 6064–6067. [Google Scholar] [CrossRef]
- Reja, S.I.; Khan, I.A.; Bhalla, V.; Kumar, M. A TICT based NIR-fluorescent probe for human serum albumin: A pre-clinical diagnosis in blood serum. Chem. Commun. 2016, 52, 1182–1185. [Google Scholar] [CrossRef]
- Kessler, M.A.; Meinitzer, A.; Petek, W.; Wolfbeis, O.S. Microalbuminuria and borderline-increased albumin excretion determined with a centrifugal analyzer and the Albumin Blue 580 fluorescence assay. Clin. Chem. 1997, 43, 996–1002. [Google Scholar] [CrossRef] [Green Version]
- Li, W.Y.; Chen, D.D.; Wang, H.; Luo, S.S.; Dong, L.C.; Zhang, Y.H.; Shi, J.B.; Tong, B.; Dong, Y.P. Quantitation of albumin in serum using “turn-on” fluorescent probe with aggregation-enhanced emission characteristics. ACS Appl. Mater. Interfaces 2015, 7, 26094–26100. [Google Scholar] [CrossRef]
- Hong, Y.; Feng, C.; Yu, Y.; Liu, J.; Lam, J.W.; Luo, K.Q.; Tang, B.Z. Quantitation, visualization, and monitoring of conformational transitions of human serum albumin by a tetraphenylethene derivative with aggregation-induced emission characteristics. Anal. Chem. 2010, 82, 7035–7043. [Google Scholar] [CrossRef]
- Min, J.; Lee, J.W.; Ahn, Y.H.; Chang, Y.T. Combinatorial dapoxyl dye library and its application to site selective probe for human serum albumin. J. Comb. Chem. 2007, 9, 1079–1083. [Google Scholar] [CrossRef]
- Jisha, V.S.; Arun, K.T.; Hariharan, M.; Ramaiah, D. Site-selective binding and dual mode recognition of serum albumin by a squaraine dye. J. Am. Chem. Soc. 2006, 128, 6024–6025. [Google Scholar] [CrossRef] [PubMed]
- Er, J.C.; Vendrell, M.; Tang, M.K.; Zhai, D.; Chang, Y.T. Fluorescent dye cocktail for multiplex drug-site mapping on human serum albumin. ACS Comb. Sci. 2013, 15, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Y.; Yu, W.T.; Hou, T.C.; Liu, T.K.; Huang, C.L.; Chen, I.C.; Tan, K.T. A selective and sensitive fluorescent albumin probe for the determination of urinary albumin. Chem. Commun. 2014, 50, 11507–11510. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Sun, W.; Wang, Z.; Peng, X.; Li, Y.; Cao, J. A fluorescent probe for site I binding and sensitive discrimination of HSA from BSA. Chem. Commun. 2014, 50, 9573–9576. [Google Scholar] [CrossRef] [PubMed]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. The characterization of two specific drug binding sites on human serum albumin. Mol. Pharmacol. 1975, 11, 824–832. [Google Scholar]
- Sudlow, G.; Birkett, D.J.; Wade, D.N.; Sudlow, G.; Birkett, D.J.; Wade, D.N. Further characterization of specific drug binding sites on human serum albumin. Mol. Pharmacol. 1976, 12, 1052–1061. [Google Scholar]
- Park, K.-S.; Yoo, K.; Kim, M.K.; Jung, W.; Choi, Y.K.; Chong, Y. A novel probe with a chlorinated α-cyanoacetophenone acceptor moiety shows near-infrared fluorescence specific for tau fibrils. Chem. Pharm. Bull. 2017, 65, 1113–1116. [Google Scholar] [CrossRef] [Green Version]
- Akman, S.; Kurt, I.; Gultepe, M.; Dibirdik, I.; Kilinc, C.; Kutluay, T.; Karaca, L.; Bingol, N.K. The development and validation of a competitive, microtiter plate enzymeimmunoassay for human albumin in urine. J. Immunoassay 1995, 16, 279–296. [Google Scholar] [CrossRef]
- Zunszain, P.A.; Ghuman, J.; Komatsu, T.; Tsuchida, E.; Curry, S. Crystal structural analysis of human serum albumin complexed with hemin and fatty acid. BMC Struct. Biol. 2003, 3. [Google Scholar] [CrossRef] [Green Version]
- Zunszain, P.A.; Ghuman, J.; McDonagh, A.F.; Curry, S. Crystallographic analysis of human serum albumin complexed with 4Z,15E-Bilirubin-IXα. Mol Biol. 2008, 381, 394–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hein, K.L.; Kragh-Hansen, U.; Morth, J.P.; Jeppesen, M.D.; Otzen, D.; Møller, J.V.; Nissen, P.J. Crystallographic analysis reveals a unique lidocaine binding site on human serum albumin. Struct. Biol. 2010, 171, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Lualdi, M.; Battaglia, L.; Colombo, A.; Leo, E.; Morelli, D.; Poiasina, E.; Vannelli, A.; Marchesini, R. Colorectal cancer detection by means of optical fluoroscopy. A study on 494 subjects. Front. Biosci. 2010, 2, 694–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwer, A.G.; Sandeep, P.M.; Goldys, E.M.; Vemulpad, S. Distinctive autofluorescence of urine samples from individuals with bacteriuria compared with normals. Clin. Chim. Acta 2009, 401, 73–75. [Google Scholar] [CrossRef]






| Name | Fluorescence properties | Sensing properties (mg/L) | Binding site d | Ref | |||
|---|---|---|---|---|---|---|---|
| λex (nm) a | λem (nm) b | Fold increase c | Limit of Detection | Detection range | |||
| TG-SA | 530 | 733 | 100 | 1.26 | 1.26–232 | I | 14 |
| ACDM | 560 | 612 | 75 | 2.5 | 0–300 | ND e | 15 |
| Indolium salt | 550 | 680 | 12 | 0.73 | 0.73–998 | I | 16 |
| AB 580 | 590 | 616 | 17 | 0.4 | 1–50 | NA f | 17 |
| DP-TPPNa | 310 | 443 | 9 | 1.68 | 1.68–100 | NA f | 18 |
| BSPOTPE | 350 | 475 | 300 | 0.67 | 0–6.7 | I/II f | 19 |
| A41-S | 360 | 473 | 55 | NA f | NA f | I | 20 |
| Squaraine Dye | 560 | 620 | 80 | NA f | NA f | II | 21 |
| BD140 | 520 | 585 | 41 | NA f | NA f | II | 22 |
| AL-1 | 456 | 490 | 400 | 0.4 | 0–66.5 | I | 23 |
| DH1 | 520 | 620 | 70 | 0.022 | 0-11.9 | I | 24 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.; Shin, E.; Jung, W.; Kim, M.K.; Chong, Y. A Near-infrared Turn-on Fluorescent Sensor for Sensitive and Specific Detection of Albumin from Urine Samples. Sensors 2020, 20, 1232. https://doi.org/10.3390/s20041232
Kim Y, Shin E, Jung W, Kim MK, Chong Y. A Near-infrared Turn-on Fluorescent Sensor for Sensitive and Specific Detection of Albumin from Urine Samples. Sensors. 2020; 20(4):1232. https://doi.org/10.3390/s20041232
Chicago/Turabian StyleKim, Yoonjeong, Eunryeol Shin, Woong Jung, Mi Kyoung Kim, and Youhoon Chong. 2020. "A Near-infrared Turn-on Fluorescent Sensor for Sensitive and Specific Detection of Albumin from Urine Samples" Sensors 20, no. 4: 1232. https://doi.org/10.3390/s20041232