Identification of Soybean Varieties Using Hyperspectral Imaging Coupled with Convolutional Neural Network
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
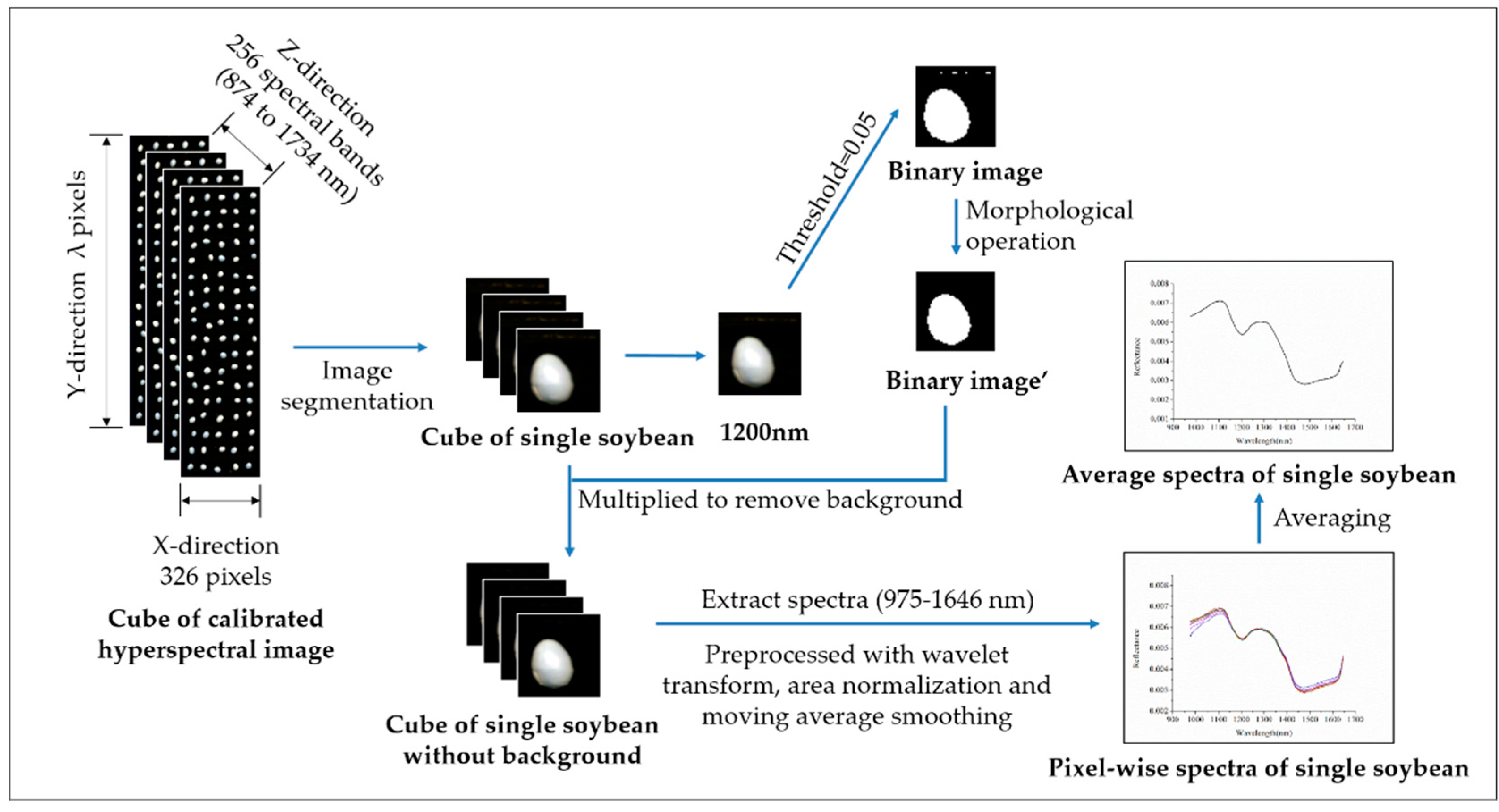
2.2. Hyperspectral Image Acquisition and Correction
2.3. Spectral Data Extraction and Preprocessing
2.4. Discrimination Models
2.4.1. Deep Learning Methods
- (1)
- Convolution layer: Used for feature learning. The kernels in convolution layers are filters with the shape of 3*1. The weights of the kernels can be automatically fitted by training. The convolution layers can recognize the patterns in spectral curves such as peaks, slopes, minimums, etc., which is similar to corners and edges in images.
- (2)
- Max pooling layer: The main features are screened out and the dimension of the feature map and calculation amount are also reduced. Thus, this layer is used to prevent over-fitting and improve the generalization ability of the model.
- (3)
- Dense layers connected with SoftMax layer: A classifier trained to establish the relationship between the extracted feature map and the corresponding classification results.
2.4.2. Principal Component Analysis
3. Results and Discussion
3.1. Spectral Profiles
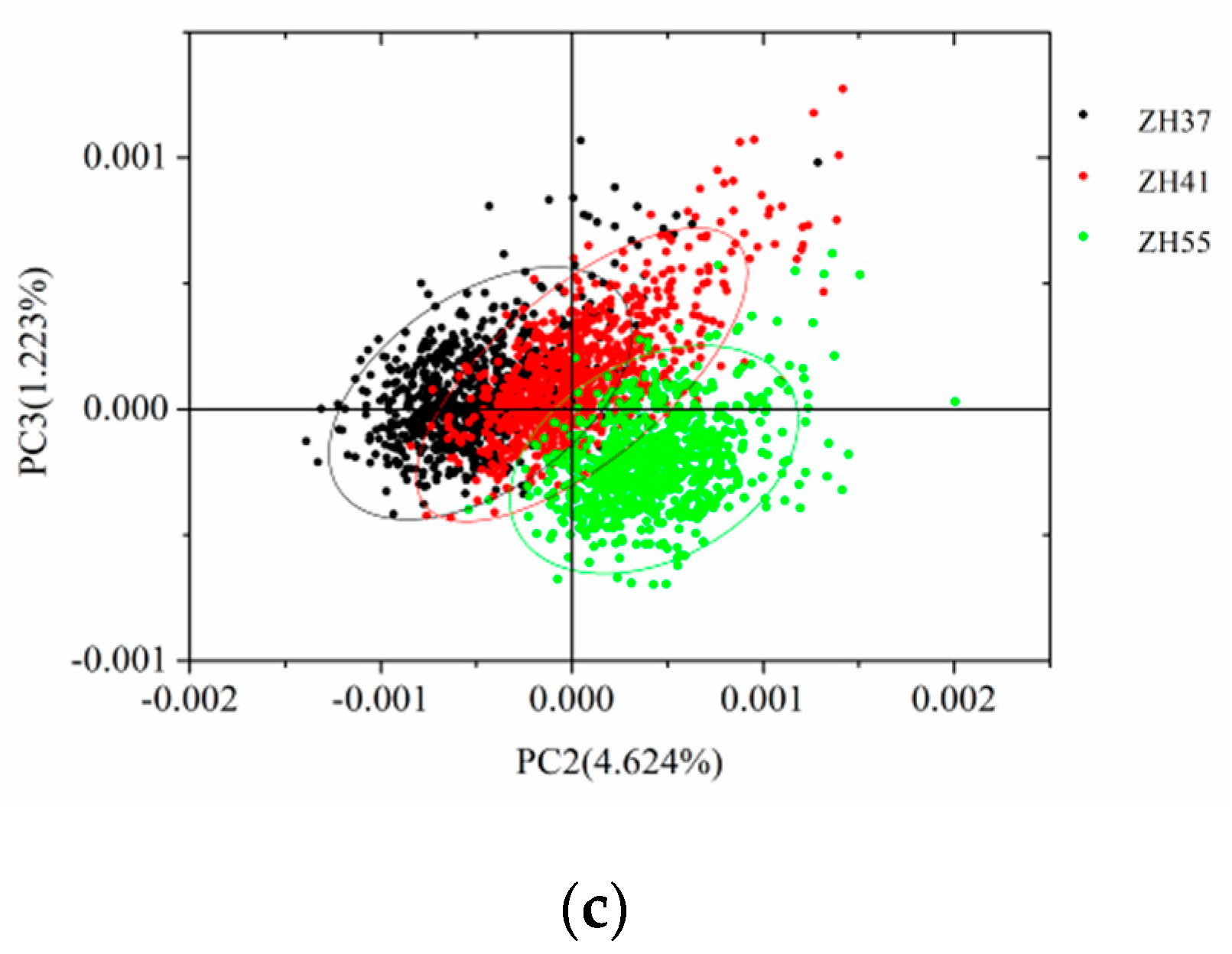
3.2. PCA Scores Scatter Plot Analysis
3.3. Classification Models on Average Spectra and Pixel-Wise Wavelengths
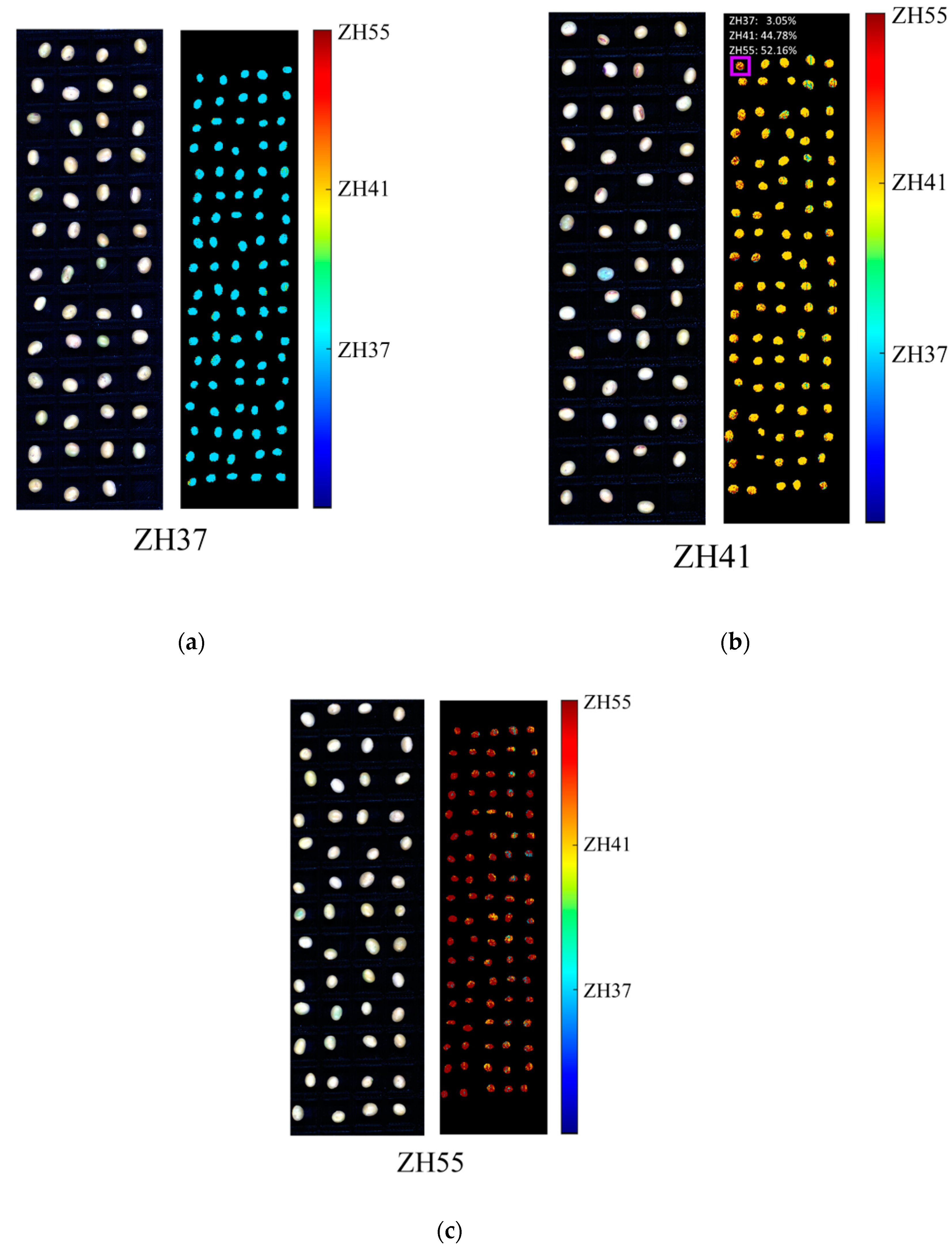
3.4. Prediction Maps
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fukushima, D. Recent progress of soybean protein foods: Chemistry, technology, and nutrition. Food Rev. Int. 1991, 7, 29. [Google Scholar] [CrossRef]
- Ribeiro, A.P.B.; Grimaldi, R.; Gioielli, L.A.; Gonçalves, L.A.G. Zero trans fats from soybean oil and fully hydrogenated soybean oil: Physico-chemical properties and food applications. Food Res. Int. 2009, 42, 401–410. [Google Scholar] [CrossRef]
- Bisen, A.; Khare, D.; Nair, P.; Tripathi, N. SSR analysis of 38 genotypes of soybean (Glycine Max (L.) Merr.) genetic diversity in India. Physiol. Mol. Biol. Plants 2015, 21, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Teng, W.; Li, W.; Zhang, Q.; Wu, D.; Zhao, X.; Li, H.; Han, Y.; Li, W. Identification of quantitative trait loci underlying seed protein content of soybean including main, epistatic, and QTL x environment effects in different regions of Northeast China. Genome 2017, 60, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Bender, R.R.; Haegele, J.W.; Below, F.E. Nutrient Uptake, Partitioning, and Remobilization in Modern Soybean Varieties. Agron. J. 2015, 107, 563–573. [Google Scholar] [CrossRef] [Green Version]
- McCarville, M.T.; Marett, C.C.; Mullaney, M.P.; Gebhart, G.D.; Tylka, G.L. Increase in Soybean Cyst Nematode Virulence and Reproduction on Resistant Soybean Varieties in Iowa From 2001 to 2015 and the Effects on Soybean Yields. Plant Health Prog. 2017, 18, 146–155. [Google Scholar] [CrossRef]
- Maria John, K.M.; Natarajan, S.; Luthria, D.L. Metabolite changes in nine different soybean varieties grown under field and greenhouse conditions. Food Chem. 2016, 211, 347–355. [Google Scholar] [CrossRef] [Green Version]
- Satturu, V.; Rani, D.; Gattu, S.; Md, J.; Mulinti, S.; Nagireddy, R.K.; Eruvuri, R.; Yanda, R. DNA Fingerprinting for Identification of Rice Varieties and Seed Genetic Purity Assessment. Agric. Res. 2018, 7, 1–12. [Google Scholar] [CrossRef]
- Tantasawat, P.; Trongchuen, J.; Prajongjai, T.; Jenweerawat, S. SSR analysis of soybean (Glycine max (L.) Merr.) genetic relationship and variety identification in Thailand. Aust. J. Crop Sci. 2011, 5, 280–287. [Google Scholar]
- Rongwen, J.; Akkaya, M.S.; Bhagwat, A.A.; Lavi, U.; Cregan, P.B. The use of microsatellite DNA markers for soybean genotype identification. Theor. Appl. Genet. 1995, 90, 43–48. [Google Scholar] [CrossRef]
- Chaugule, A.A.; Mali, S.N. Identification of paddy varieties based on novel seed angle features. Comput. Electron. Agric. 2016, 123, 415–422. [Google Scholar] [CrossRef]
- Kiratiratanapruk, K.; Sinthupinyo, W. Color and texture for corn seed classification by machine vision. In Proceedings of the 2011 International Symposium on Intelligent Signal Processing and Communications Systems (ISPACS), Chiang Mai, Thailand, 7–9 December 2011; pp. 1–5. [Google Scholar]
- Jacobsen, S.; Søndergaard, I.; Møller, B.; Desler, T.; Munck, L. A chemometric evaluation of the underlying physical and chemical patterns that support near infrared spectroscopy of barley seeds as a tool for explorative classification of endosperm genes and gene combinations. J. Cereal Sci. 2005, 42, 281–299. [Google Scholar] [CrossRef]
- Seregély, Z.; Deák, T.; Bisztray, G.D. Distinguishing melon genotypes using NIR spectroscopy. Chemom. Intell. Lab. Syst. 2004, 72, 195–203. [Google Scholar] [CrossRef]
- Wu, D.; Sun, D.W. Advanced applications of hyperspectral imaging technology for food quality and safety analysis and assessment: A review - Part I: Fundamentals. Innovative Food Sci. Emerg. Technol. 2013, 19, 1–14. [Google Scholar] [CrossRef]
- Gowen, A.A.; O’Donnell, C.P.; Cullen, P.J.; Downey, G.; Frias, J.M. Hyperspectral imaging—An emerging process analytical tool for food quality and safety control. Trends Food Sci. Technol. 2007, 18, 590–598. [Google Scholar] [CrossRef]
- Lorente, D.; Aleixos, N.; Gomez-Sanchis, J.; Cubero, S.; Garcia-Navarrete, O.L.; Blasco, J. Recent Advances and Applications of Hyperspectral Imaging for Fruit and Vegetable Quality Assessment. Food Bioprocess Technol. 2012, 5, 1121–1142. [Google Scholar] [CrossRef]
- Feng, L.; Zhu, S.; Liu, F.; He, Y.; Bao, Y.; Zhang, C. Hyperspectral imaging for seed quality and safety inspection: A review. Plant Methods 2019, 15, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Jian, C.; Zhao, Y.; Zhu, S.; Yong, H.; Chu, Z. Variety Identification of Single Rice Seed Using Hyperspectral Imaging Combined with Convolutional Neural Network. Appl. Sci. 2018, 8, 212. [Google Scholar] [CrossRef]
- Soares, S.F.C.; Medeiros, E.P.; Pasquini, C.; Morello, C.D.L.; Galvão, R.K.H.; Araújo, M.C.U. Classification of individual cotton seeds with respect to variety using near-infrared hyperspectral imaging. Anal. Methods 2016, 8, 8498–8505. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, F.; He, Y. Identification of coffee bean varieties using hyperspectral imaging: influence of preprocessing methods and pixel-wise spectra analysis. Sci.Rep. 2018, 8, 2166. [Google Scholar] [CrossRef]
- Lara, M.A.; Lleo, L.; Diezma-Iglesias, B.; Roger, J.M.; Ruiz-Altisent, M. Monitoring spinach shelf-life with hyperspectral image through packaging films. J. Food Eng. 2013, 119, 353–361. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Li, C.; Yang, F. Classification of foreign matter embedded inside cotton lint using short wave infrared (SWIR) hyperspectral transmittance imaging. Comput. Electron. Agric. 2017, 139, 75–90. [Google Scholar] [CrossRef]
- Acquarelli, J.; van Laarhoven, T.; Gerretzen, J.; Tran, T.N.; Buydens, L.M.C.; Marchiori, E. Convolutional neural networks for vibrational spectroscopic data analysis. Anal. Chim. Acta 2017, 954, 22–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Lin, T.; Xu, J.; Luo, X.; Ying, Y. DeepSpectra: An end-to-end deep learning approach for quantitative spectral analysis. Anal. Chim. Acta 2019, 1058, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ye, Y.; Li, X.; Lau, R.Y.K.; Zhang, X.; Huang, X. Hyperspectral Image Classification With Deep Learning Models. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5408–5423. [Google Scholar] [CrossRef]
- Yu, X.; Tang, L.; Wu, X.; Lu, H. Nondestructive Freshness Discriminating of Shrimp Using Visible/Near-Infrared Hyperspectral Imaging Technique and Deep Learning Algorithm. Food Anal. Methods 2018, 11, 768–780. [Google Scholar] [CrossRef]
- Yue, J.; Mao, S.; Li, M. A deep learning framework for hyperspectral image classification using spatial pyramid pooling. Remote Sens. Lett. 2016, 7, 875–884. [Google Scholar] [CrossRef]
- Signoroni, A.; Savardi, M.; Baronio, A.; Benini, S. Deep Learning Meets Hyperspectral Image Analysis: A Multidisciplinary Review. J. Imaging 2019, 5, 52. [Google Scholar] [CrossRef]
- Yu, X.; Lu, H.; Liu, Q. Deep-learning-based regression model and hyperspectral imaging for rapid detection of nitrogen concentration in oilseed rape (Brassica napus L.) leaf. Chemom. Intell. Lab. Syst. 2018, 172, 188–193. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, S.; Zhang, C.; Feng, X.; Feng, L.; He, Y. Application of hyperspectral imaging and chemometrics for variety classification of maize seeds. RSC Adv. 2018, 8, 1337–1345. [Google Scholar] [CrossRef] [Green Version]
- Feng, L.; Zhu, S.; Zhou, L.; Zhao, Y.; Bao, Y.; Zhang, C.; He, Y. Detection of Subtle Bruises on Winter Jujube Using Hyperspectral Imaging With Pixel-Wise Deep Learning Method. IEEE Access 2019, 7, 64494–64505. [Google Scholar] [CrossRef]
- Feng, L.; Zhu, S.; Zhang, C.; Bao, Y.; Gao, P.; He, Y. Variety Identification of Raisins Using Near-Infrared Hyperspectral Imaging. Molecules 2018, 23, 2907. [Google Scholar] [CrossRef]
- Chen, X.; Xie, L.; He, Y.; Guan, T.; Zhou, X.; Wang, B.; Feng, G.; Yu, H.; Ji, Y. Fast and accurate decoding of Raman spectra-encoded suspension arrays using deep learning. Analyst 2019, 144, 4312–4319. [Google Scholar] [CrossRef]




| Component | Near-Infrared Hyperspectral Imaging System |
|---|---|
| Imaging spectrograph | ImSpector N17E (Spectral Imaging Ltd., Oulu, Finland) |
| Camera | InGaAs camera (Xeva 992; Xenics Infrared Solutions, Leuven, Belgium) |
| Lens | OLES22 (Spectral Imaging Ltd., Oulu, Finland) |
| Image size (Image width × image length × wavebands) | 326 × λ × 256 |
| Acquisition mode | Line-scan |
| Light sources | 3900 Lightsource (Illumination Technologies Inc., Syracuse, New York, USA) |
| Mobile platform | IRCP0076 electric displacement table (Isuzu Optics Corp., Taiwan) |
| Number 1 | Accuracy (%) | Computation Time 5 (s) | ||
|---|---|---|---|---|
| Tra-average 2 | Val-average 3 | Pre-average 4 | ||
| 10 | 100 | 75.556 | 87.296 | 5.742 |
| 20 | 100 | 79.259 | 88.593 | 6.103 |
| 30 | 100 | 85.370 | 93.370 | 6.490 |
| 60 | 100 | 90.370 | 96.333 | 7.016 |
| 90 | 100 | 94.074 | 97.778 | 11.590 |
| 180 | 100 | 95.185 | 98.222 | 15.862 |
| 360 | 100 | 98.333 | 99.259 | 21.662 |
| 540 | 100 | 99.074 | 99.296 | 28.260 |
| 720 | 100 | 99.259 | 99.481 | 35.667 |
| 810 | 100 | 99.444 | 99.778 | 38.935 |
| Model | Number 1 | Accuracy (%) | Computation Time 5 (s) | ||
|---|---|---|---|---|---|
| Tra-average 2 | Val-average 3 | Pre-average 4 | |||
| ResNet | 10 | 100 | 61.111 | 74.000 | 18.210 |
| 810 | 100 | 93.333 | 97.556 | 190.509 | |
| Inception | 10 | 100 | 74.630 | 89.111 | 50.004 |
| 810 | 100 | 96.852 | 98.889 | 95.604 | |
| Number 1. | Pixels 2 | Accuracy (%) | Computaion Time 7 (s) | |||||
|---|---|---|---|---|---|---|---|---|
| Training | Validation | Prediction | Tra-pixel 3 | Val-pixel 4 | Pre-pixel 5 | Pre-average 6 | ||
| 10 | 12,546 | 208,788 | 1,057,007 | 94.165 | 73.002 | 74.741 | 79.741 | 315 |
| 20 | 24,719 | 93.337 | 73.632 | 74.736 | 80.370 | 468 | ||
| 30 | 37,280 | 94.612 | 76.102 | 77.864 | 88.556 | 558 | ||
| 60 | 75,969 | 92.069 | 81.725 | 83.875 | 95.556 | 1140 | ||
| Set | Number 1 | Accuracy (%) | |||
|---|---|---|---|---|---|
| ZH37 | ZH41 | ZH55 | All | ||
| Tra-pixel 2 | 10 | 99.604 | 94.138 | 88.035 | 94.165 |
| 20 | 99.429 | 92.553 | 87.214 | 93.337 | |
| 30 | 98.989 | 94.390 | 89.876 | 94.612 | |
| 60 | 97.056 | 83.849 | 94.552 | 92.069 | |
| Val-pixel 3 | 10 | 91.677 | 76.386 | 49.875 | 73.002 |
| 20 | 90.519 | 79.339 | 50.071 | 73.632 | |
| 30 | 86.811 | 82.075 | 58.803 | 76.102 | |
| 60 | 82.521 | 78.030 | 84.583 | 81.725 | |
| Pre-pixel 4 | 10 | 94.938 | 77.774 | 51.766 | 74.741 |
| 20 | 94.703 | 78.737 | 51.069 | 74.736 | |
| 30 | 92.277 | 82.186 | 59.416 | 77.864 | |
| 60 | 87.901 | 79.681 | 83.859 | 83.875 | |
| Tra-vote 5 | 10 | 100 | 100 | 100 | 100 |
| 20 | 100 | 100 | 100 | 100 | |
| 30 | 100 | 100 | 100 | 100 | |
| 60 | 100 | 100 | 100 | 100 | |
| Val-vote 6 | 10 | 100 | 96.111 | 58.333 | 84.815 |
| 20 | 100 | 98.333 | 58.333 | 85.556 | |
| 30 | 100 | 98.889 | 72.222 | 90.370 | |
| 60 | 99.444 | 96.111 | 98.889 | 98.148 | |
| Pre-vote 7 | 10 | 100 | 96.111 | 57.889 | 84.667 |
| 20 | 100 | 97.000 | 57.556 | 84.852 | |
| 30 | 100 | 99.222 | 74.444 | 91.222 | |
| 60 | 100 | 96.667 | 99.667 | 98.778 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, S.; Zhou, L.; Zhang, C.; Bao, Y.; Wu, B.; Chu, H.; Yu, Y.; He, Y.; Feng, L. Identification of Soybean Varieties Using Hyperspectral Imaging Coupled with Convolutional Neural Network. Sensors 2019, 19, 4065. https://doi.org/10.3390/s19194065
Zhu S, Zhou L, Zhang C, Bao Y, Wu B, Chu H, Yu Y, He Y, Feng L. Identification of Soybean Varieties Using Hyperspectral Imaging Coupled with Convolutional Neural Network. Sensors. 2019; 19(19):4065. https://doi.org/10.3390/s19194065
Chicago/Turabian StyleZhu, Susu, Lei Zhou, Chu Zhang, Yidan Bao, Baohua Wu, Hangjian Chu, Yue Yu, Yong He, and Lei Feng. 2019. "Identification of Soybean Varieties Using Hyperspectral Imaging Coupled with Convolutional Neural Network" Sensors 19, no. 19: 4065. https://doi.org/10.3390/s19194065





