Multivariate Analysis of Difference Raman Spectra of the Irradiated Nucleus and Cytoplasm Region of SH-SY5Y Human Neuroblastoma Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Treatment
2.2. Raman Micro-Spectroscopy Measurements
2.3. Data Analysis
2.3.1. Preliminary Analysis
2.3.2. Difference Spectra and interval-Principal Component Analysis (i-PCA)
3. Results and Discussion
3.1. Analysis of Nucleus- and Cytoplasm-Related Spectra
3.2. Multivariate Analysis for Spectral Interval Selection
3.3. Analysis of Difference Spectra
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef] [PubMed]
- Ember, K.J.I.; Hoeve, M.A.; McAughtrie, S.L.; Bergholt, M.S.; Dwyer, B.J.; Stevens, M.M.; Faulds, K.; Forbes, S.J.; Campbell, C.J. Raman spectroscopy and regenerative medicine: A review. NPJ Regen. Med. 2017, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Shipp, D.W.; Sinjab, F.; Notingher, I. Raman spectroscopy: Techniques and applications in the life sciences. Adv. Opt. Photon 2017, 9, 315. [Google Scholar] [CrossRef]
- Kuhar, N.; Sil, S.; Verma, T.; Umapathy, S. Challenges in application of Raman spectroscopy to biology and materials. RSC Adv. 2018, 8, 25888–25908. [Google Scholar] [CrossRef]
- Cordero, E.; Latka, I.; Matthaus, C.; Schie, I.W.; Popp, J. In-vivo Raman spectroscopy: From basics to applications. J. Biomed. Opt. 2018, 23, 071210. [Google Scholar] [CrossRef] [PubMed]
- Camerlingo, C.; Verde, A.; Manti, L.; Meschini, R.; Delfino, I.; Lepore, M. Graphene-Based Raman Spectroscopy for pH Sensing of X-rays Exposed and Unexposed Culture Media and Cells. Sensors 2018, 18, 2242. [Google Scholar] [CrossRef] [PubMed]
- D’Apuzzo, F.; Perillo, L.; Delfino, I.; Portaccio, M. Monitoring early phases of orthodontic treatment by means of Raman spectroscopies. J. Biomed. Opt. 2017, 22, 1. [Google Scholar] [CrossRef] [PubMed]
- Kann, B.; Offerhaus, H.L.; Windbergs, M.; Otto, C. Raman microscopy for cellular investigations—From single cell imaging to drug carrier uptake visualization. Adv. Drug Deliv. Rev. 2015, 89, 71–90. [Google Scholar] [CrossRef]
- Byrne, H.J.; Bonnier, F.; Casey, A.; Maher, M.; McIntyre, J.; Efeoglu, E.; Farhane, Z. Advancing Raman microspectroscopy for cellular and subcellular analysis: towards in vitro high-content spectralomic analysis. Appl. Opt. 2018, 57, E11–E19. [Google Scholar] [CrossRef]
- Bonnier, F.; Byrne, H.J. Understanding the molecular information contained in principal component analysis of vibrational spectra of biological systems. Analyst 2012, 137, 322–332. [Google Scholar] [CrossRef]
- Lieber, C.A.; Mahadevan-Jansen, A. Automated method for subtraction of fluorescence from biological Raman spectra. Appl. Spectrosc. 2003, 57, 1363–1367. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, E.B.; Manoharan, R.; Koo, T.-W.; E Shafer, K.; Motz, J.T.; Fitzmaurice, M.; Kramer, J.R.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Prospects for in vivo Raman spectroscopy. Phys. Med. Biol. 2000, 45, R1–R59. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, J.; Huang, X.; Li, G.; Liu, B. Goldindec: A Novel Algorithm for Raman Spectrum Baseline Correction. Appl. Spectrosc. 2015, 69, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Gallo, C.; Capozzi, V.; Lasalvia, M.; Perna, G. An algorithm for estimation of background signal of Raman spectra from biological cell samples using polynomial functions of different degrees. Vib. Spectrosc. 2016, 83, 132–137. [Google Scholar] [CrossRef]
- Varmuza, K.; Filzmoser, P. Introduction to Multivariate Statistical Analysis in Chemometrics; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2009; p. 321. [Google Scholar]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Tech. Instrum. 2015, 2. art. n. 8 (38 pages). [Google Scholar] [CrossRef]
- Matthews, Q.; Brolo, A.; Lum, J.; Duan, X.; Jirasek, A. Raman spectroscopy of single human tumour cells exposed to ionizing radiation in vivo. Phys. Med. Biol. 2011, 56, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Harder, S.J.; Matthews, Q.; Isabelle, M.; Brolo, A.G.; Lum, J.J.; Jirasek, A. A Raman Spectroscopic Study of Cell Response to Clinical Doses of Ionizing Radiation. Appl. Spectrosc. 2015, 69, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Delfino, I.; Perna, G.; Lasalvia, M.; Capozzi, V.; Manti, L.; Camerlingo, C.; Lepore, M. Visible micro-Raman spectroscopy of single human mammary epithelial cells exposed to x-ray radiation. J. Biomed. Opt. 2015, 20, 35003. [Google Scholar] [CrossRef]
- Delfino, I.; Perna, G.; Ricciardi, V.; Lasalvia, M.; Manti, L.; Capozzi, V.; Lepore, M. X-ray irradiation effects on nuclear and membrane regions of single SH-SY5Y human neuroblastoma cells investigated by Raman micro-spectroscopy. J. Pharm. Biomed. Anal. 2019, 164, 557–573. [Google Scholar] [CrossRef]
- Van Nest, S.J.; Nicholson, L.M.; DeVorkin, L.; Brolo, A.G.; Lum, J.J.; Jirasek, A. Raman Spectroscopic Signatures Reveal Distinct Biochemical and Temporal Changes in Irradiated Human Breast Adenocarcinoma Xenografts. Radiat. Res. 2018, 189, 497–504. [Google Scholar] [CrossRef]
- Meksiarun, P.; Aoki, P.H.B.; Van Nest, S.J.; Sobral-Filho, R.G.; Lum, J.J.; Brolo, A.G.; Jirasek, A. Breast cancer subtype specific biochemical responses to radiation. Analyst 2018, 143, 3850–3858. [Google Scholar] [CrossRef] [PubMed]
- Meade, A.D.; Howe, O.; Unterreiner, V.; Sockalingum, G.D.; Byrne, H.J.; Lyng, F.M. Vibrational spectroscopy in sensing radiobiological effects: analyses of targeted and non-targeted effects in human keratinocytes. Faraday Discuss. 2016, 187, 213–234. [Google Scholar] [CrossRef] [PubMed]
- Harder, S.J.; Isabelle, M.; DeVorkin, L.; Smazynski, J.; Beckham, W.; Brolo, A.G.; Lum, J.J.; Jirasek, A. Raman spectroscopy identifies radiation response in human non-small cell lung cancer xenografts. Sci. Rep. 2016, 6, 21006. [Google Scholar] [CrossRef] [PubMed]
- Lasalvia, M.; Perna, G.; Manti, L.; Rasero, J.; Stramaglia, S.; Capozzi, V. Raman spectroscopy monitoring of MCF10A cells irradiated by protons at clinical doses. Int. J. Radiat. Boil. 2019, 95, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Lasalavia, M.; Perna, G.; Pisciotta, P.; Cammarata, F.P.; Manti, L.; Capozzi, V. Raman spectroscopy for the evaluation of the radiobiological sensitivity of normal human breast cells at different time points after irradiation by a clinical proton beam. Analyst 2019, 144, 2097–2108. [Google Scholar] [CrossRef] [PubMed]
- Baskar, R.; Dai, J.; Wenlong, N.; Yeo, R.; Yeoh, K.-W. Biological response of cancer cells to radiation treatment. Front. Mol. Biosci. 2014, 1, 24. [Google Scholar] [CrossRef]
- Harvey, T.J.; Hughes, C.; Ward, A.D.; Faria, E.C.; Henderson, A.; Clarke, N.W.; Brown, M.D.; Snook, R.D.; Gardner, P. Classification of fixed urological cells using Raman tweezers. J. Biophotonics 2009, 2, 47–69. [Google Scholar] [CrossRef]
- Meade, A.D.; Clarke, C.; Draux, F.; Sockalingum, G.D.; Manfait, M.; Lyng, F.M.; Byrne, H.J. Studies of chemical fixation effects in human cell lines using Raman microspectroscopy. Anal. Bioanal. Chem. 2010, 396, 1781–1791. [Google Scholar] [CrossRef]
- Maguire, A.; Vegacarrascal, I.; White, L.; McClean, B.; Howe, O.; Lyng, F.M.; Meade, A.D. Analyses of Ionizing Radiation Effects In Vitro in Peripheral Blood Lymphocytes with Raman. Spectrosc. Radiat. Res. 2015, 183, 407–416. [Google Scholar] [CrossRef]
- Meade, A.D.; Maguire, A.; Bryant, J.; Cullen, D.; Medipally, D.; White, L.; McClean, B.; Shields, L.; Armstrong, J.; Dunne, M.; et al. Prediction of DNA damage and G2 chromosomal radio-sensitivity ex vivo in peripheral blood mononuclear cells with label-free Raman micro-spectroscopy. Int. J. Radiat. Biol. 2019, 95, 44–53. [Google Scholar] [CrossRef]
- Lasch, P. Spectral pre-processing for biomedical vibrational spectroscopy and micro-spectroscopic imaging. Chemometr. Intell. Lab. Syst. 2013, 117, 100–114. [Google Scholar] [CrossRef]
- Leardi, R.; Nørgaard, L. Sequential application of backward interval partial least squares and genetic algorithms for the selection of relevant spectral regions. J. Chemom. 2004, 18, 486–497. [Google Scholar] [CrossRef]
- Goodpaster, A.M.; Kennedy, M.A. Quantification and statistical significance analysis of group separation in NMR-based metabolomics studies. Chemom. Intell. Lab. Syst. 2011, 109, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Ong, Y.H.; Lim, M.; Liu, Q. Comparison of principal component analysis and biochemical component analysis in Raman spectroscopy for the discrimination of apoptosis and necrosis in K562 leukemia cells. Opt. Express 2012, 20, 22158–22171. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Gestl, E.; Eckert, K.; Allara, D.; Irudayaraj, J. Characterization of human breast epithelial cells by confocal Raman microspectroscopy. Cancer Detect. Prev. 2006, 30, 515–522. [Google Scholar] [CrossRef]
- Notingher, I.; Verrier, S.; Romanska, H.; Bishop, A.E.; Polak, J.M.; Hench, L.L. In situ Characterisation of Living Cells by Raman Spectroscopy. Spectrosc. 2002, 16, 43–51. [Google Scholar] [CrossRef]
- Schulze, H.G.; Konorov, S.O.; Aparicio, S.A.; Piret, J.M.; Blades, M.W.; Turner, R.F.B. Raman Microscopy of Human Embryonic Stem Cells Exposed to Heat and Cold Stress. Appl. Spectrosc. 2011, 65, 1380–1386. [Google Scholar] [CrossRef]
- Kumar, S.; Verma, T.; Mukherjee, R.; Ariese, F.; Somasundaram, K.; Umapathy, S. Raman and infra-red microspectroscopy: Towards quantitative evaluation for clinical research by ratiometric analysis. Chem. Soc. Rev. 2016, 45, 1879–1900. [Google Scholar] [CrossRef]
- Kim, S.; Seo, M.; Oh, J.M.; Cho, E.A.; Juhnn, Y.S. Inhibition of gamma ray-induced apoptosis by stimulatory heterotrimeric GTP binding protein involves Bcl-xL down-regulation in SH-SY5Y human neuroblastoma cells. Exp. Mol. Med. 2007, 39, 583–593. [Google Scholar] [CrossRef][Green Version]
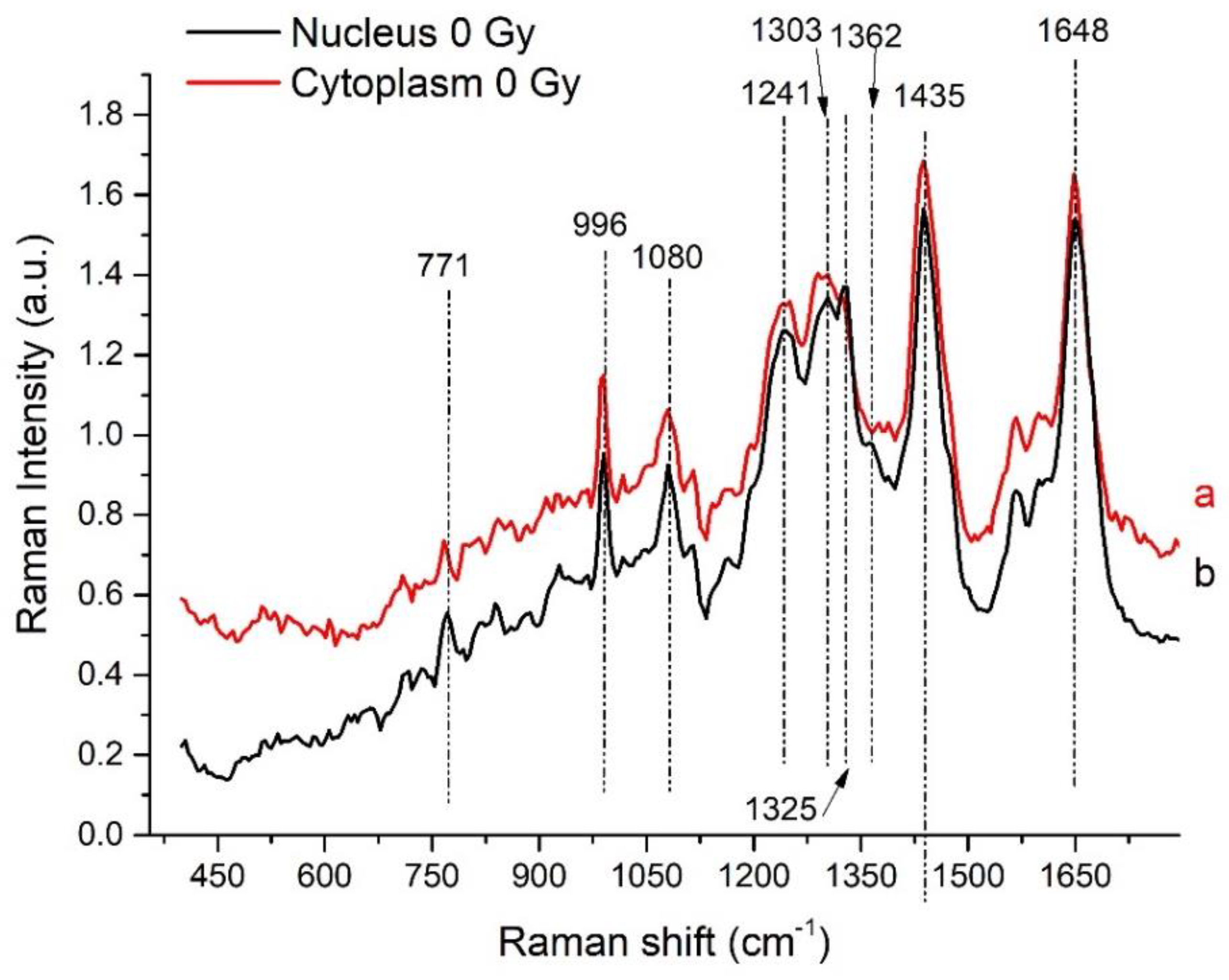

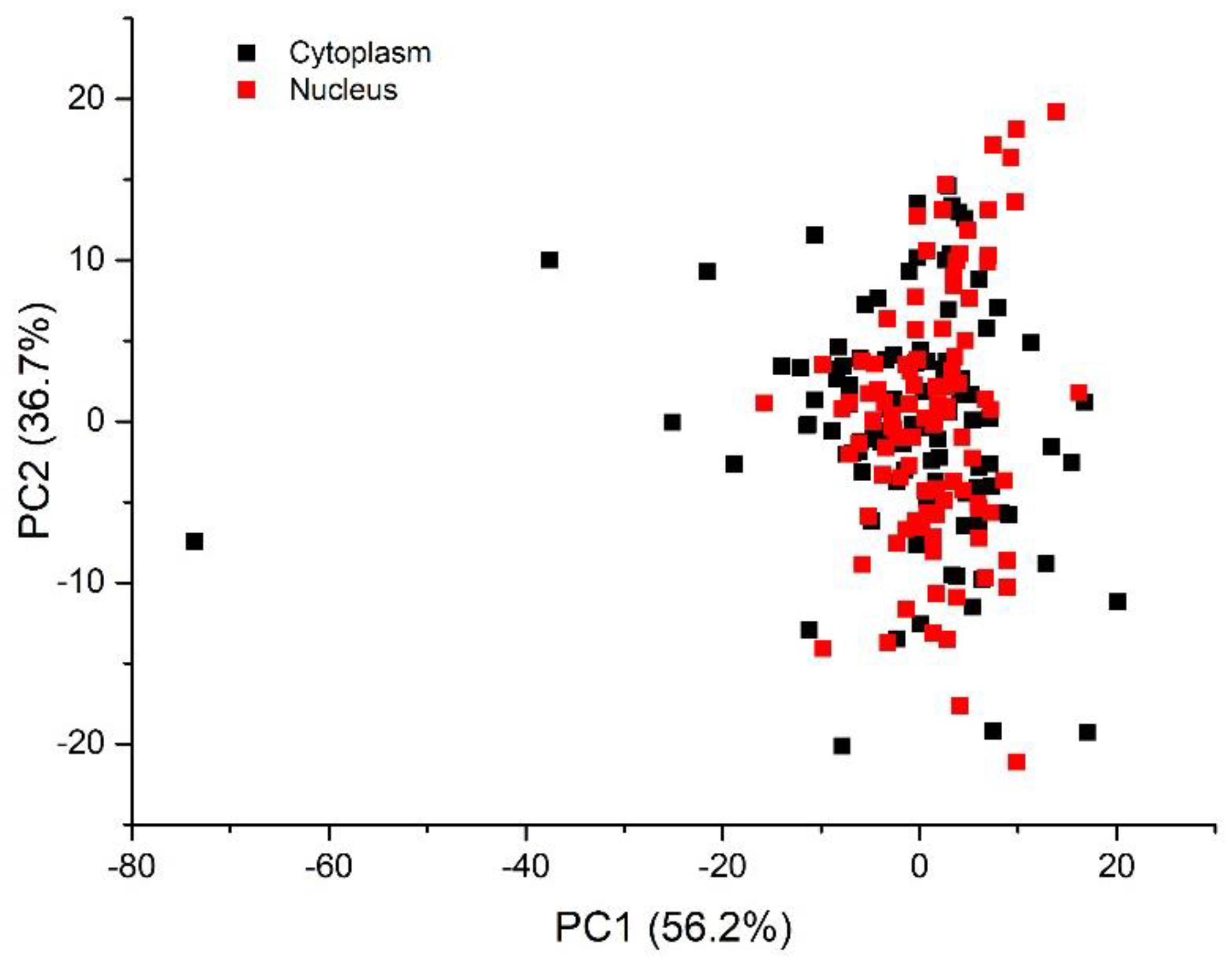
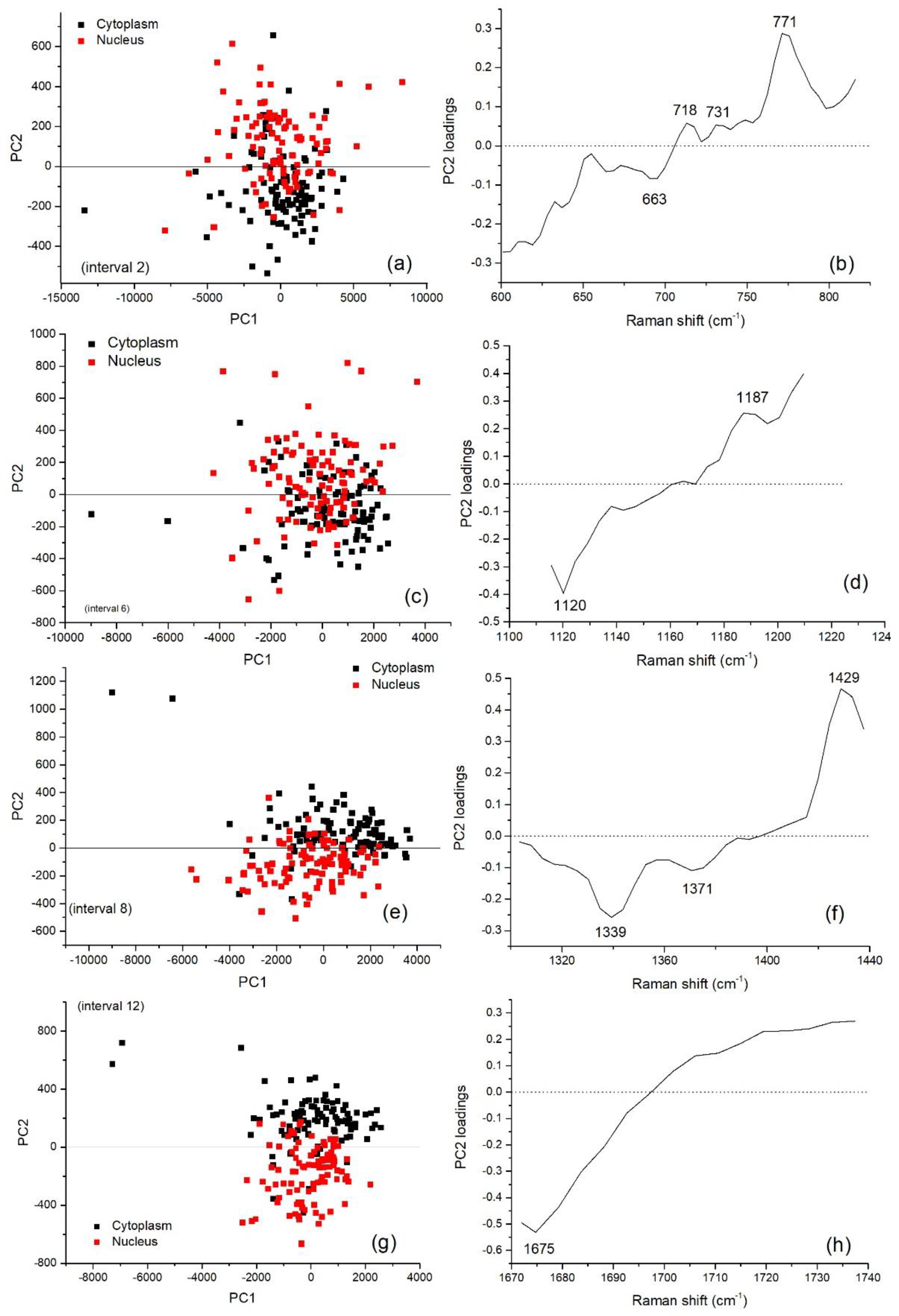
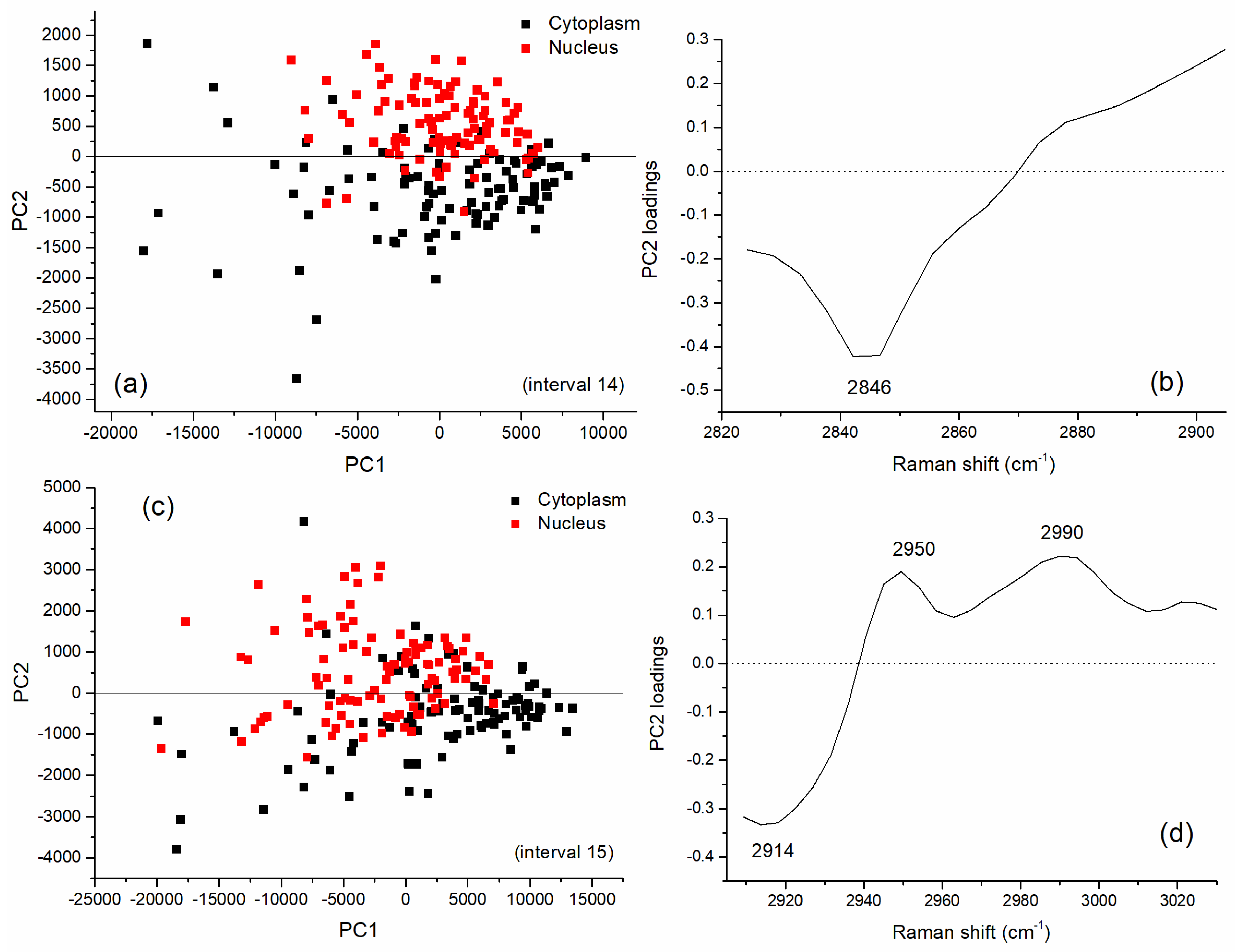


| Peak (cm−1) | DNA/RNA | Protein | Lipid/Carbohydrate |
|---|---|---|---|
| 3057 | Aromatics | Aromatics | |
| 2994–3008 | CH str. | ||
| 2950–2980 | CH3 asym. str. | CH3 asym str | |
| 2918–2940 | CH2 asym. str. | CH2 asym. str. | |
| 2885 | CH3 sym. str. | ||
| 2860 | CH2 sym. str. | ||
| 1640–1670 | Amide I (α-helix, β-sheet, coil) | C = C sym. str. | |
| 1621 | Tyrosine/Tryptophan C = C | ||
| 1594–1608 | Phenylalanine/Tyrosine C = C | ||
| 1572–1581 | Adenine, Guanine | ||
| 1487 | Adenine, Guanine | ||
| 1447–1460 | CH2/CH3 ben., CH def. | CH2 sc., CH def. | |
| 1421–1430 | Adenine, Guanine, CH2 bk. | ||
| 1374 | Thymine | ||
| 1338–1345 | Adenine, Guanine | CH def. | |
| 1326 | Guanine | CH def. | |
| 1309 | Adenine | Amide III | CH2 tw. |
| 1297 | CH2 tw. | ||
| 1256–1263 | Amide III (α-helix) | CH2 def. | |
| 1248–1255 | Amide III (β-sheets) | ||
| 1219–1235 | Amide III (random coil) | ||
| 1190–1204 | Tryptophan/Tyrosine/Phenylalanine CH sym. str., CH ben. | ||
| 1168–1178 | Phenylalanine/Tyrosine, CH ben. | ||
| 1123 | CN sym. str. | CC asym. str., CO str. | |
| 1090–1098 | O-P-O str. | CC chain sym. str., CO, C = C str. | |
| 1051–1066 | CO sym. str. | CC chain sym. str., CO, C = C str. | |
| 1020–1032 | Phenylalanine, CH in-p. ben. | ||
| 996–1003 | Phenylalanine sym. ring br. | ||
| 973–986 | PO23- sym. str. | CC sym. str., β-sheets | CH ben. |
| 955–963 | CH3 def. | ||
| 932–938 | CC sym. str., α-helix | COC glycos. bond | |
| 883 | cDNA | CCN+ sym. str., COC ring | |
| 870 | C1 - C2 str. | ||
| 849–853 | Tyrosine ring br. Proline CC str. | ||
| 823–840 | O-P-O bk. | Tyrosine ring br. | |
| 807–811 | O-P-O str. RNA | O-P-O str. | |
| 770–785 | Cytosine/Thymine/Uracil ring br. O-P-O str. | ||
| 753–767 | Tryptophan ring br. | ||
| 722–728 | Adenine | Choline | |
| 704 | Cholesterol | ||
| 677 | Thymine, Guanine | ||
| 655 | Tyrosine (skeletal) |
| Peak (cm−1) | Assignment | Dependence upon Increasing Dose |
|---|---|---|
| 3003 | l/c (CH str.) | Contribution decreases |
| 2955 | p and l/c (CH3 asym. str.) | Contribution increases |
| 2940 | p and l/c (CH2 asym. str.) | Contribution decreases |
| 2860 | l/c (CH2 sym. str.) | Contribution decreases |
| 1640–1670 | p (Amide I -α-helix, β-sheet, coil) and l/c (C = C sym. str.) | 1640 cm−1 contribution increases; 1661 cm−1 contribution decreases |
| 1621 | p (Tyrosine/Tryptophan C = C) | Contribution decreases |
| 1430 | d (Adenine, Guanine, CH2 bk.) | Contribution decreases |
| 1374 | d (Thymine) | Contribution increases |
| 1344 | d (Adenine, Guanine) | Contribution increases |
| 1320–1330 | d (Adenine, Guanine, CH2 bk.) | Contribution decreases |
| 807 | d (O-P-O str. RNA) | Contribution decreases |
| 780 | d (Cytosine/Thymine/Uracil ring br., O-P-O str.) | Contribution decreases |
| 767 | p (Tryptophan ring br.) | Contribution decreases |
| 722 | d (Adenine) and l/c (Choline) | Contribution increases |
| 704 | l/c (Cholesterol) | Contribution increases |
| 677 | d (Thymine, Guanine) | Contribution increases |
| 655 | p (Tyrosine skeletal) | Contribution increases |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delfino, I.; Ricciardi, V.; Manti, L.; Lasalvia, M.; Lepore, M. Multivariate Analysis of Difference Raman Spectra of the Irradiated Nucleus and Cytoplasm Region of SH-SY5Y Human Neuroblastoma Cells. Sensors 2019, 19, 3971. https://doi.org/10.3390/s19183971
Delfino I, Ricciardi V, Manti L, Lasalvia M, Lepore M. Multivariate Analysis of Difference Raman Spectra of the Irradiated Nucleus and Cytoplasm Region of SH-SY5Y Human Neuroblastoma Cells. Sensors. 2019; 19(18):3971. https://doi.org/10.3390/s19183971
Chicago/Turabian StyleDelfino, Ines, Valerio Ricciardi, Lorenzo Manti, Maria Lasalvia, and Maria Lepore. 2019. "Multivariate Analysis of Difference Raman Spectra of the Irradiated Nucleus and Cytoplasm Region of SH-SY5Y Human Neuroblastoma Cells" Sensors 19, no. 18: 3971. https://doi.org/10.3390/s19183971
APA StyleDelfino, I., Ricciardi, V., Manti, L., Lasalvia, M., & Lepore, M. (2019). Multivariate Analysis of Difference Raman Spectra of the Irradiated Nucleus and Cytoplasm Region of SH-SY5Y Human Neuroblastoma Cells. Sensors, 19(18), 3971. https://doi.org/10.3390/s19183971









