DNA Biosensor Based on Double-Layer Discharge for the Detection of HPV Type 16
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. DNA Probe Immobilization and Hybridization with DNA Target
2.3. Electrochemical Measurements
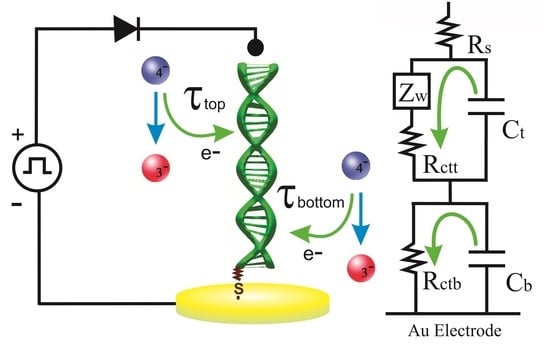
2.4. The Potential Relaxation Method
3. Results and Discussion
3.1. Step Potential
3.2. Potential Relaxation Curves and DNA Equivalent Circuit Model
3.3. Analytical Performance of the DNA Biosensor
3.4. Specificity of the DNA Biosensor
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Shah, S.S.; Senapati, S.; Klacsmann, F.; Miller, D.L.; Johnson, J.J.; Chang, H.-C.; Stack, M.S. Current Technologies and Recent Developments for Screening of HPV-Associated Cervical and Oropharyngeal Cancers. Cancers 2016, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kumar, P.; Das, B.C. HPV: Molecular pathways and targets. Curr. Probl. Cancer 2018, 42, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Brotherton, J.M.L.; Tabrizi, S.N.; Phillips, S.; Pyman, J.; Cornall, A.M.; Lambie, N.; Anderson, L.; Cummings, M.; Payton, D.; Scurry, J.P.; et al. Looking beyond human papillomavirus (HPV) genotype 16 and 18: Defining HPV genotype distribution in cervical cancers in Australia prior to vaccination. Int. J. Cancer 2017, 141, 1576–1584. [Google Scholar] [CrossRef] [PubMed]
- Philp, L.; Jembere, N.; Wang, L.; Gao, J.; Maguire, B.; Kupets, R. Pap tests in the diagnosis of cervical cancer: Help or hinder? Gynecol. Oncol. 2018, 150, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Parmin, N.A.; Hashim, U.; Gopinath, S.C.B. Designing probe from E6 genome region of human Papillomavirus 16 for sensing applications. Int. J. Biol. Macromol. 2018, 107, 1738–1746. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, M.; Liu, X.; Sharma, A.; Ding, X. A Point-of-Need infrared mediated PCR platform with compatible lateral flow strip for HPV detection. Biosens. Bioelectron. 2017, 96, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.-H.; Kim, D.-E.; Im, J.-H.; Kang, S.-J.; Lee, D.-H.; Son, S.J. Rapid visual identification of PCR amplified nucleic acids by centrifugal gel separation: Potential use for molecular point-of-care tests. Biosens. Bioelectron. 2016, 79, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Serour, Y.; Bendahmane, M.; Abbou Baker, F.; Medles, M.; Moueddene, B.; Kraiba, R. HPV test by Hybrid Capture II for the diagnosis of HR-HPV persistent infection. Médecine Et Mal. Infect. 2017, 47, 484–489. [Google Scholar] [CrossRef]
- Cook, D.A.; Smith, L.W.; Law, J.; Mei, W.; van Niekerk, D.J.; Ceballos, K.; Gondara, L.; Franco, E.L.; Coldman, A.J.; Ogilvie, G.S.; et al. Aptima HPV Assay versus Hybrid Capture® 2 HPV test for primary cervical cancer screening in the HPV FOCAL trial. J. Clin. Virol. 2017, 87, 23–29. [Google Scholar] [CrossRef]
- Frías, I.A.; Avelino, K.Y.; Silva, R.R.; Andrade, C.A.; Oliveira, M.D. Trends in Biosensors for HPV: Identification and Diagnosis. J. Sens. 2015, 2015, 16. [Google Scholar] [CrossRef]
- Jearanaikoon, P.; Prakrankamanant, P.; Leelayuwat, C.; Wanram, S.; Limpaiboon, T.; Promptmas, C. The evaluation of loop-mediated isothermal amplification-quartz crystal microbalance (LAMP-QCM) biosensor as a real-time measurement of HPV16 DNA. J. Virol. Methods 2016, 229, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, A.M.J.; Moulick, A.; Richtera, L.; Krejcova, L.; Kalina, L.; Datta, R.; Svobodova, M.; Hynek, D.; Masarik, M.; Heger, Z.; et al. Dual-color quantum dots-based simultaneous detection of HPV-HIV co-infection. Sens. Actuators B Chem. 2018, 258, 295–303. [Google Scholar] [CrossRef]
- Inada, N.M.; Buzza, H.H.; Carbinatto, F.M.; Blanco, K.C.; de Andrade, C.T.; Vollet-Filho, J.D.; Bagnato, V.S.; Allison, R.R. Optical techniques for the diagnosis and treatment of lesions induced by the human papillomavirus—A resource letter. Photodiagn. Photodyn. Ther. 2018, 23, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Ponzo, I.; Möller, F.M.; Daub, H.; Matscheko, N. A DNA-Based Biosensor Assay for the Kinetic Characterization of Ion-Dependent Aptamer Folding and Protein Binding. Molecules 2019, 24, 2877. [Google Scholar] [CrossRef] [PubMed]
- Chekin, F.; Bagga, K.; Subramanian, P.; Jijie, R.; Singh, S.K.; Kurungot, S.; Boukherroub, R.; Szunerits, S. Nucleic aptamer modified porous reduced graphene oxide/MoS2 based electrodes for viral detection: Application to human papillomavirus (HPV). Sens. Actuators B Chem. 2018, 262, 991–1000. [Google Scholar] [CrossRef]
- Lu, W.; Yuan, Q.; Yang, Z.; Yao, B. Self-primed isothermal amplification for genomic DNA detection of human papillomavirus. Biosens. Bioelectron. 2017, 90, 258–263. [Google Scholar] [CrossRef]
- Jampasa, S.; Siangproh, W.; Laocharoensuk, R.; Yanatatsaneejit, P.; Vilaivan, T.; Chailapakul, O. A new DNA sensor design for the simultaneous detection of HPV type 16 and 18 DNA. Sens. Actuators B Chem. 2018, 265, 514–521. [Google Scholar] [CrossRef]
- Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Molecular Biosensors for Electrochemical Detection of Infectious Pathogens in Liquid Biopsies: Current Trends and Challenges. Sensors 2017, 17, 2533. [Google Scholar] [CrossRef]
- Xu, Q.; Davis, J.J. The Diagnostic Utility of Electrochemical Impedance. Electroanalysis 2014, 26, 1249–1258. [Google Scholar] [CrossRef]
- Uygun, Z.O.; Ertuǧrul Uygun, H.D. A short footnote: Circuit design for faradaic impedimetric sensors and biosensors. Sens. Actuators B Chem. 2014, 202, 448–453. [Google Scholar] [CrossRef]
- Li, X.; Ahmadi, M.; Collins, L.; Kalinin, S.V. Deconvolving distribution of relaxation times, resistances and inductance from electrochemical impedance spectroscopy via statistical model selection: Exploiting structural-sparsity regularization and data-driven parameter tuning. Electrochim. Acta 2019, 313, 570–583. [Google Scholar] [CrossRef]
- Shariati, M.; Ghorbani, M.; Sasanpour, P.; Karimizefreh, A. An ultrasensitive label free human papilloma virus DNA biosensor using gold nanotubes based on nanoporous polycarbonate in electrical alignment. Anal. Chim. Acta 2018. [Google Scholar] [CrossRef] [PubMed]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Henry, C.S.; Vilaivan, T.; Chailapakul, O. Electrochemical paper-based peptide nucleic acid biosensor for detecting human papillomavirus. Anal. Chim. Acta 2017, 952, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Karimizefreh, A.; Mahyari, F.A.; VaezJalali, M.; Mohammadpour, R.; Sasanpour, P. Impedimetic biosensor for the DNA of the human papilloma virus based on the use of gold nanosheets. Mikrochim. Acta 2017, 184, 1729–1737. [Google Scholar] [CrossRef]
- Dharuman, V.; Nebling, E.; Grunwald, T.; Albers, J.; Blohm, L.; Elsholz, B.; Wörl, R.; Hintsche, R. DNA hybridization detection on electrical microarrays using coulostatic pulse technique. Biosens. Bioelectron. 2006, 22, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Keighley, S.D.; Li, P.; Estrela, P.; Migliorato, P. Optimization of DNA immobilization on gold electrodes for label-free detection by electrochemical impedance spectroscopy. Biosens. Bioelectron. 2008, 23, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Balaguera, E.; Polo, J.L. A generalized procedure for the coulostatic method using a constant phase element. Electrochim. Acta 2017, 233, 167–172. [Google Scholar] [CrossRef]
- Sun, A.C.; Alvarez-Fontecilla, E.; Venkatesh, A.G.; Aronoff-Spencer, E.; Hall, D.A. High-Density Redox Amplified Coulostatic Discharge-Based Biosensor Array. IEEE J. Solid-State Circuits 2018, 53, 2054–2064. [Google Scholar] [CrossRef] [PubMed]
- Nassi, A.; Guillon, F.X.; Amar, A.; Hainque, B.; Amriche, S.; Maugé, D.; Markova, E.; Tsé, C.; Bigey, P.; Lazerges, M.; et al. Electrochemical DNA-biosensors based on long-range electron transfer: Optimization of the amperometric detection in the femtomolar range using two-electrode setup and ultramicroelectrode. Electrochim. Acta 2016, 209, 269–277. [Google Scholar] [CrossRef]
- Bajwa, N.; Maldonado, C.J.; Thundat, T.; Passian, A. Piezoresistive measurement of Swine H1N1 Hemagglutinin peptide binding with microcantilever arrays. AIP Adv. 2014, 4, 037118. [Google Scholar] [CrossRef]
- Kim, S.; Yi, D.; Passian, A.; Thundat, T. Observation of an anomalous mass effect in microcantilever-based biosensing caused by adsorbed DNA. Appl. Phys. Lett. 2010, 96, 153703. [Google Scholar] [CrossRef]









| Oligonucleotide | Sequence |
|---|---|
| Probe | 5′-HS(CH2)6GTCATTATGTGCTGCCATATCTACTT-CAGA-3’ |
| Complementary | 5´-TCTGAAGTAGATATGGCAGCACATAATGAC-3´, |
| Single-base mismatch | 5´ TCTGAAATAGATATGGCAGCACATAATGAC-3´ |
| Parameter | ssDNA Value | ssDNA-dsDNA Difference Δ % |
|---|---|---|
| −178.06 ± 0.19 | 1.76 ± 0.1 | |
| 16.86 ± 0.25 | 31.85 ± 1.15 | |
| 2.82 ± 0.33 | 78.72 ± 2.83 | |
| 10.43 ± 0.15 | 25.02 ± 1.13 | |
| 0.909 ± 0.01 | 18.81 ± 1.61 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Espinosa, J.R.; Galván, M.; Quiñones, A.S.; Ayala, J.L.; Durón, S.M. DNA Biosensor Based on Double-Layer Discharge for the Detection of HPV Type 16. Sensors 2019, 19, 3956. https://doi.org/10.3390/s19183956
Espinosa JR, Galván M, Quiñones AS, Ayala JL, Durón SM. DNA Biosensor Based on Double-Layer Discharge for the Detection of HPV Type 16. Sensors. 2019; 19(18):3956. https://doi.org/10.3390/s19183956
Chicago/Turabian StyleEspinosa, José R., Marisol Galván, Arturo S. Quiñones, Jorge L. Ayala, and Sergio M. Durón. 2019. "DNA Biosensor Based on Double-Layer Discharge for the Detection of HPV Type 16" Sensors 19, no. 18: 3956. https://doi.org/10.3390/s19183956