Towards Non-Invasive Methods to Assess Population Structure and Biomass in Vulnerable Sea Pen Fields †
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
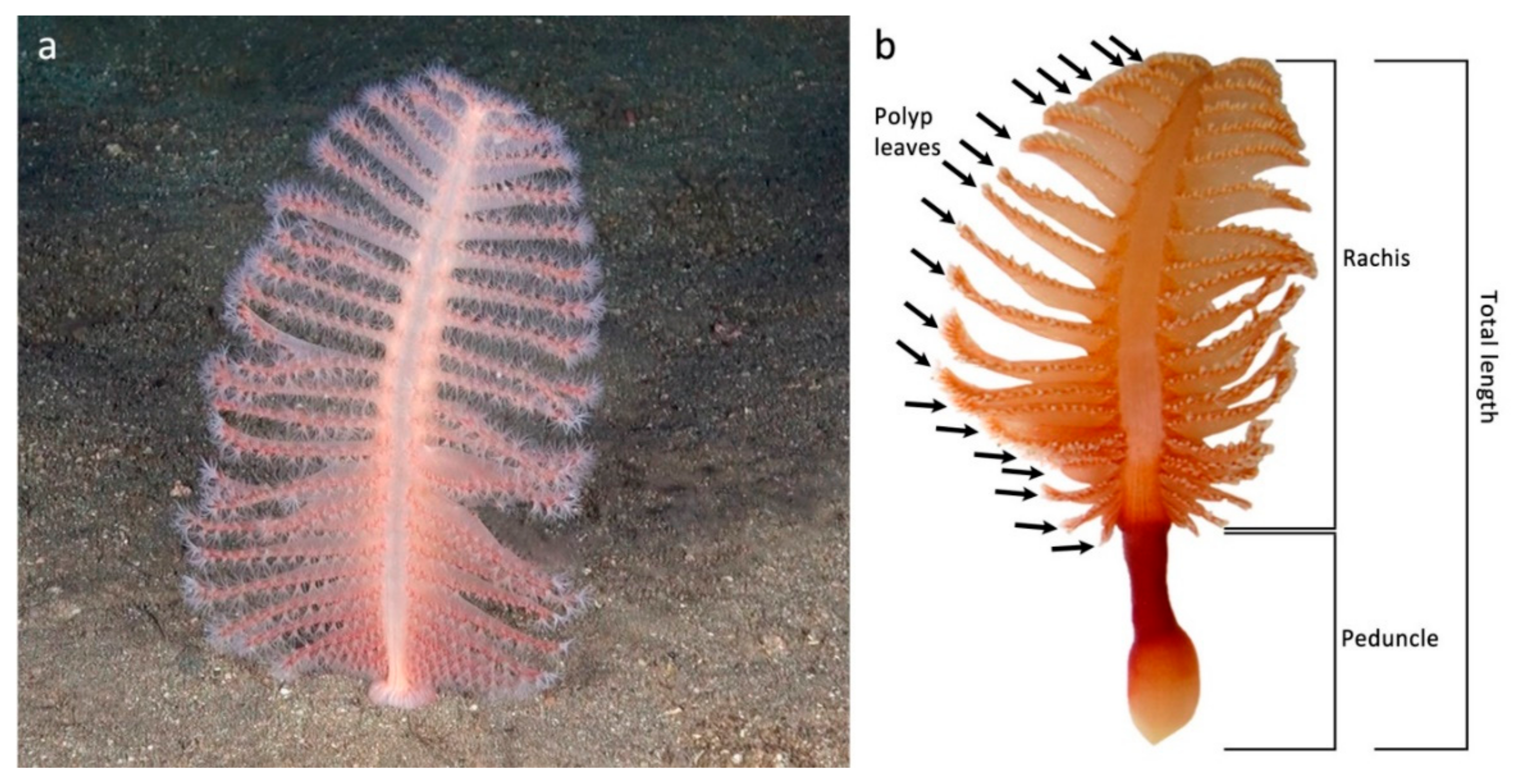
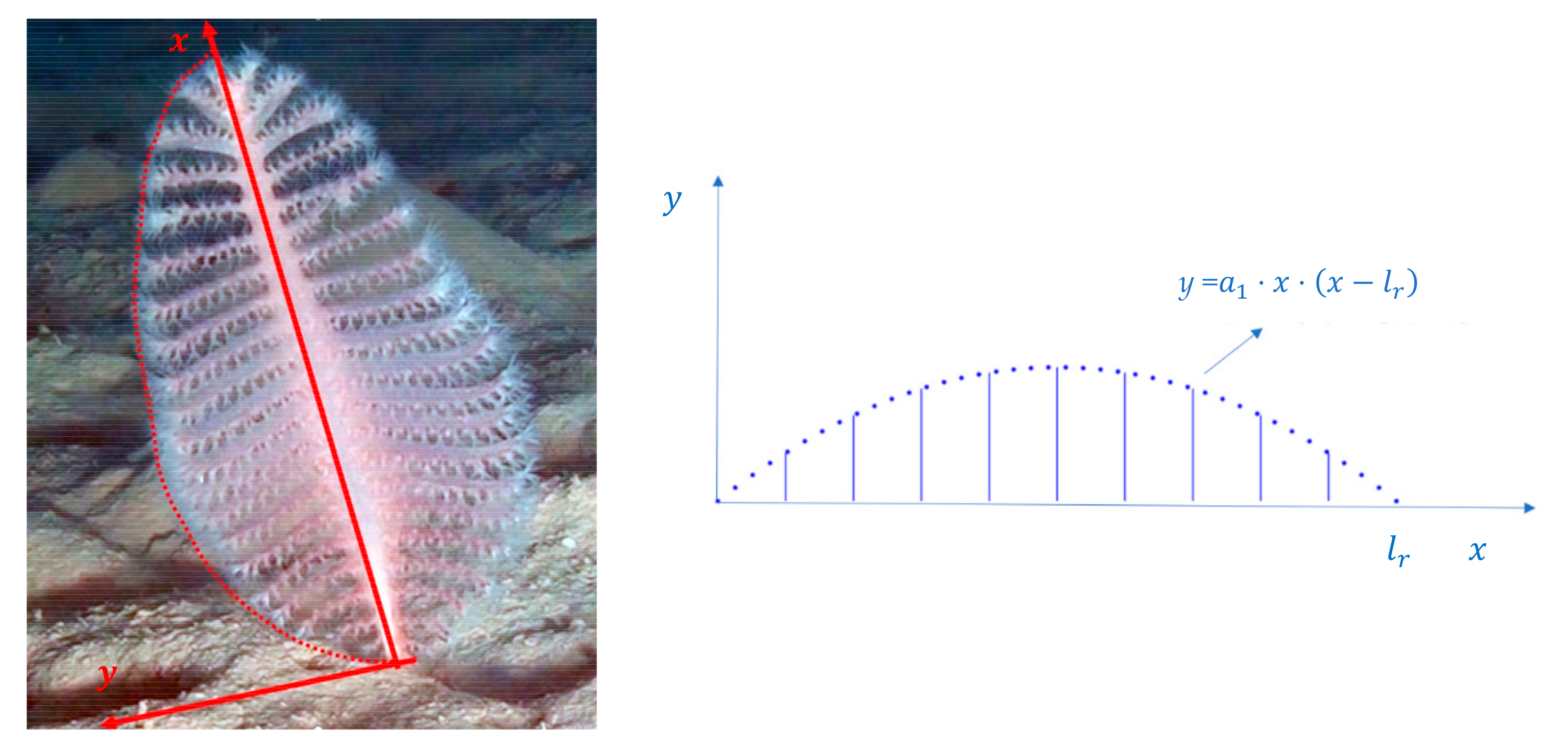
3.1. Model for Size Estimation
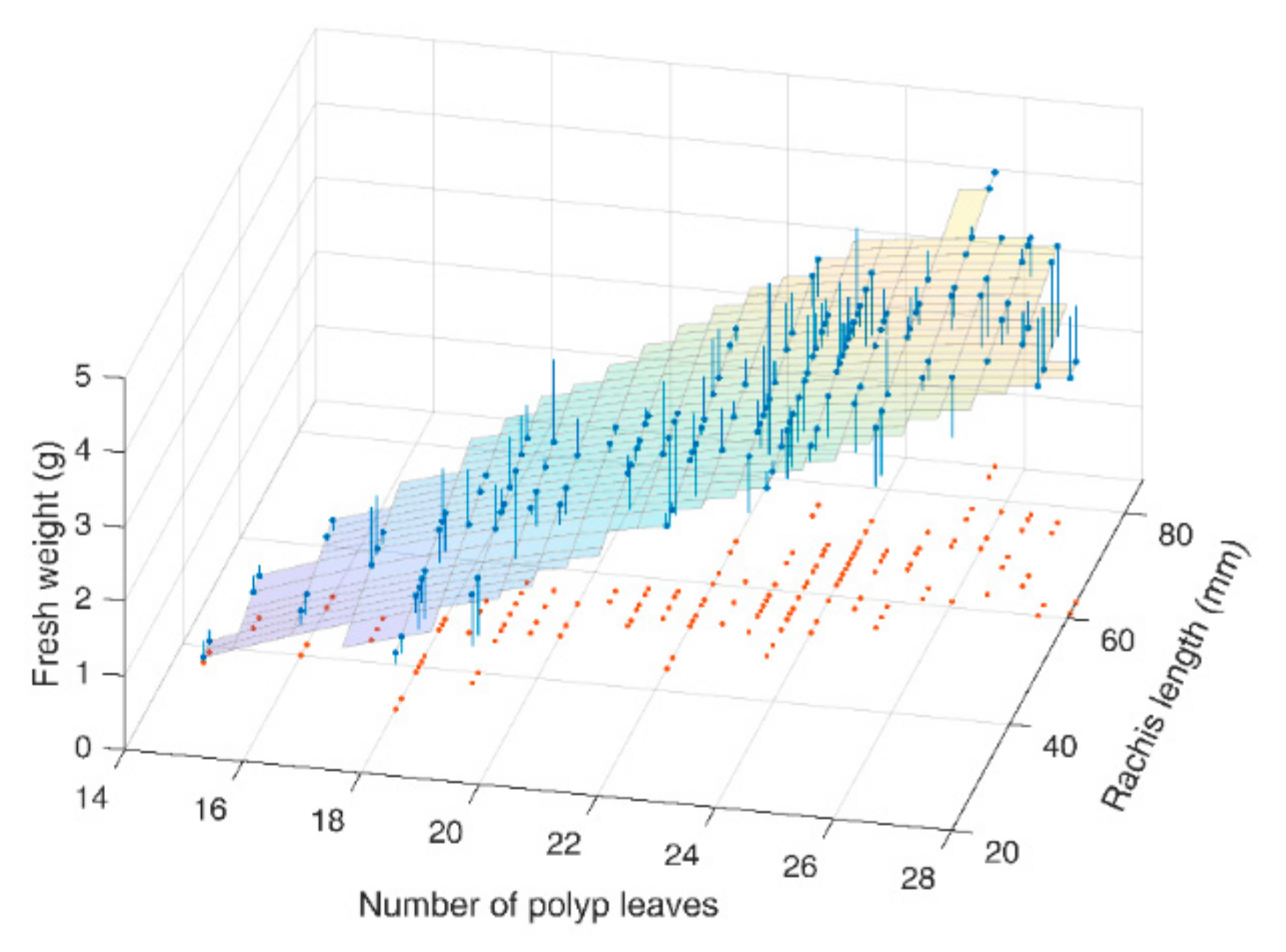
3.2. Model for Fresh Weight Estimation
3.3. Dry Weight Estimation
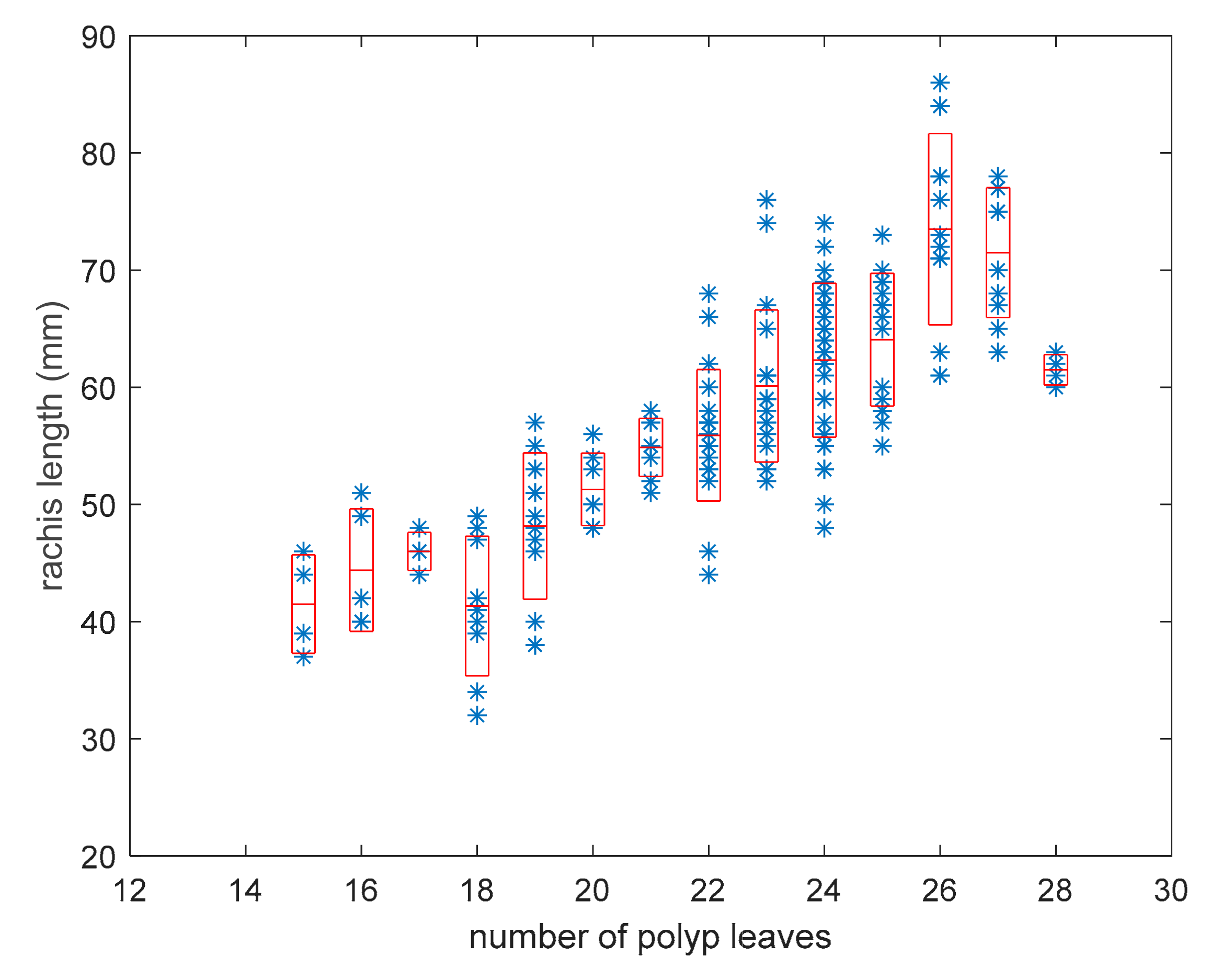
3.4. Distribution of the Number of Polyp Leaves
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chimienti, G.; Bo, M.; Mastrototaro, F. Know the distribution to assess the changes: Mediterranean cold-water coral bioconstructions. Rendiconti Lincei Scienze Fisiche e Naturali 2018, 29, 583–588. [Google Scholar] [CrossRef]
- Chimienti, G.; Bo, M.; Taviani, M.; Mastrototaro, F. Occurrence and Biogeography of Mediterranean Cold-Water Corals. In Mediterranean Cold-Water Corals: Past, Present and Future; Orejas, C., Jimennez, C., Eds.; Springer International Publishing: Berlin, Germany, 2019; Volume 9, pp. 1–64. [Google Scholar]
- Rossi, S.; Bramanti, L.; Gori, A.; Orejas, C. An overview of the animal forests of the world. In Marine Animal Forests; Rossi, S., Ed.; Springer International Publishing: Berlin, Germany, 2017; pp. 1–26. [Google Scholar]
- Mastrototaro, F.; Chimienti, G.; Acosta, J.; Blanco, J.; Garcia, S.; Rivera, J.; Aguilar, R. Isidella elongata (Cnidaria: Alcyonacea) facies in the western Mediterranean Sea: Visual surveys and descriptions of its ecological role. Eur. Zool. J. 2017, 84, 209–225. [Google Scholar] [CrossRef]
- Porporato, E.M.D.; De Domenico, F.; Mangano, M.C.; Spanò, N. Macropodia longirostris and Latreillia elegans (Decapoda, Brachyura) climbing on Mediterranean Pennatulidae (Anthozoa, Octocorallia): A preliminary note. Crustaceana 2011, 84, 1777–1780. [Google Scholar]
- Baillon, S.; Hamel, J.F.; Wareham, V.E.; Mercier, A. Deep cold-water corals as nurseries for fish larvae. Front. Ecol. Environ. 2012, 10, 351–356. [Google Scholar] [CrossRef]
- Kenchington, E.; Murillo, F.J.; Lirette, C.; Sacau, M.; Koen-Alonso, M.; Kenny, A.; Ollerhead, N.; Wareham, V.; Beazley, L. Kernel Density Surface Modelling as a Means to Identify Significant Concentrations of Vulnerable Marine Ecosystem Indicators. PLoS ONE 2014, 9, e109365. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Pico, S.; Serrano, A.; Punzón, A.; Altuna, A.; Fernández-Zapico, O.; Velasco, F. Sea pen (Pennatulacea) aggregations on the northern Spanish shelf: Distribution and faunal assemblages. Sci. Mar. 2017, 81, 1–11. [Google Scholar] [CrossRef]
- FAO (Food and Agriculture Organization). International Guidelines for the Management of Deep-Sea Fisheries in the High Seas; FAO: Rome, Italy, 2009; pp. 1–21. [Google Scholar]
- Chimienti, G.; Angeletti, L.; Rizzo, L.; Tursi, A.; Mastrototaro, F. ROV vs. trawling approaches in the study of benthic communities: The case of Pennatula rubra (Cnidaria: Cennatulacea). J. Mar. Biol. Assoc. U. K. 2018, 98, 1859–1869. [Google Scholar] [CrossRef]
- Adamo, F.; Attivissimo, F.; Di Nisio, A.; Savino, M. An automated visual inspection system for the glass Industry. In Proceedings of the 16th IMEKO TC-4 International Symposium and 13th Workshop on ADC Modelling and Testing, Florence, Italy, 22–24 September 2008; pp. 442–447. [Google Scholar]
- Adamo, F.; Attivissimo, F.; Di Nisio, A. Calibration of an inspection system for online quality control of satin glass. IEEE Trans. Instrum. Meas. 2010, 59, 1035–1046. [Google Scholar] [CrossRef]
- Attivissimo, F.; Di Nisio, A.; Guarnieri Calò Carducci, C.; Spadavecchia, M. Fast Thermal Characterization of Thermoelectric Modules Using Infrared Camera. IEEE Trans. Instrum. Meas. 2017, 66, 305–314. [Google Scholar] [CrossRef]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef]
- Sharma, N.; Aggarwal, L.M. Automated medical image segmentation techniques. J. Med. Phys. 2010, 35, 3–14. [Google Scholar] [CrossRef]
- Adamo, F.; Attivissimo, F.; Di Nisio, A.; Spadavecchia, M. An automatic document processing system for medical data extraction. Measurement 2015, 61, 88–99. [Google Scholar] [CrossRef]
- Andria, G.; Attivissimo, F.; Di Nisio, A.; Lanzolla, A.M.L.; Maiorana, A.; Mangiatini, M.; Spadavecchia, M. Dosimetric Characterization and Image Quality Assessment in Breast Tomosynthesis. IEEE Trans. Instrum. Meas. 2017, 66, 2535–2544. [Google Scholar] [CrossRef]
- Gleason, A.C.R.; Reid, R.P.; Voss, K.J. Automated classification of underwater multispectral imagery for coral reef monitoring. In Proceedings of the OCEANS 2007, Vancouver, BC, Canada, 29 September–4 October 2007; pp. 1–8. [Google Scholar]
- Phinn, S.R.; Roelfsema, C.M.; Mumby, P.J. Multi-scale, object-based image analysis for mapping geomorphic and ecological zones on coral reefs. Int. J. Remote Sens. 2012, 33, 3768–3797. [Google Scholar] [CrossRef]
- Foglini, F.; Grande, V.; Marchese, F.; Bracchi, V.A.; Prampolini, M.; Angeletti, L.; Castellan, G.; Chimienti, G.; Hansen, I.M.; Gudmundsen, M.; et al. Underwater Hyperspectral Imaging for seafloor and benthic habitat mapping in the southern Adriatic Sea (Italy). In Proceedings of the 2018 IEEE International Workshop on Metrology for the Sea, Learning to Measure Sea Health Parameters (MetroSea), Bari, Italy, 8–10 October 2018; pp. 201–205. [Google Scholar]
- Murillo, F.J.; MacDonald, B.W.; Kenchington, E.; Campana, S.E.; Sainte-Marie, B.; Sacau, M. Morphometry and growth of sea pen species from dense habitats in the Gulf of St. Lawrence, eastern Canada. Mar. Biol. Res. 2018, 14, 366–382. [Google Scholar] [CrossRef]
- Ludvigsen, M.; Johnsen, G.; Sørensen, A.J.; Lågstad, P.A.; Ødegård, Ø. Scientific operations combining ROV and AUV in the Trondheim Fjord. Mar. Technol. Soc. J. 2014, 48, 59–71. [Google Scholar] [CrossRef]
- Mastrototaro, F.; Aguilar, R.; Chimienti, G.; Gravili, C.; Boero, F. The rediscovery of Rosalinda incrustans (Cnidaria: Hydrozoa) in the Mediterranean Sea. Ital. J. Zool. 2016, 83, 244–247. [Google Scholar] [CrossRef]
- Petritoli, E.; Leccese, F.; Cagnetti, M. A High Accuracy Buoyancy System Control for an Underwater Glider. In Proceedings of the 2018 IEEE International Workshop on Metrology for the Sea, Learning to Measure Sea Health Parameters (MetroSea), Bari, Italy, 8–10 October 2018; pp. 257–261. [Google Scholar]
- Adamo, F.; Andria, G.; Di Nisio, A.; Calò Carducci, C.G.; Lay-Ekuakille, A.; Mattencini, G.; Spadavecchia, M. Designing and prototyping a sensors head for test and certification of UAV components. Int. J. Smart Sens. Intell. Syst. 2017, 10, 646–672. [Google Scholar] [CrossRef]
- Barwise, A.; Cowie, M. Advancement in Geotechnical Site Investigation Practice Using ROV Technology. In Proceedings of the Offshore Technology Conference, Houston, TX, USA, 6–9 May 2013; pp. 1–8. [Google Scholar]
- McLean, D.L.; Macreadie, P.; White, D.J.; Thomson, P.G.; Fowler, A.; Gates, A.R.; Benfield, M.; Horton, T.; Skropeta, D.; Bond, T.; et al. Understanding the Global Scientific Value of Industry ROV Data, to Quantify Marine Ecology and Guide Offshore Decommissioning Strategies. In Proceedings of the Offshore Technology Conference Asia, Kuala Lumpur, Malaysia, 20–23 March 2018; pp. 1–10. [Google Scholar]
- Adamo, F.; Andria, G.; Bottiglieri, O.; Cotecchia, F.; Di Nisio, A.; Miccoli, D.; Sollecito, F.; Spadavecchia, M.; Todaro, F.; Trotta, A.; et al. GeoLab, a measurement system for the geotechnical characterization of polluted submarine sediments. Measurement 2018, 127, 335–347. [Google Scholar] [CrossRef]
- Chimienti, G.; Tursi, A.; Mastrototaro, F. Biometric relationships in the red sea pen Pennatula rubra (Cnidaria: Pennatulacea). In Proceedings of the 2018 IEEE International Workshop on Metrology for the Sea, Learning to Measure Sea Health Parameters (MetroSea), Bari, Italy, 8–10 October 2018; pp. 212–216. [Google Scholar]
- Rooper, C.N.; Wilborn, P.; Goddard, P.; Williams, K.; Towler, R.; Hoff, G.R. Validation of deep-sea coral and sponge distribution models in the Aleutian Islands, Alaska. ICES J. Mar. Sci. 2017, 75, 199–209. [Google Scholar] [CrossRef]
- Chimienti, G.; Angeletti, L.; Mastrototaro, F. Withdrawal behaviour of the red sea pen Pennatula rubra (Cnidaria: Pennatulacea). Eur. Zool. J. 2018, 85, 64–70. [Google Scholar] [CrossRef]
- Otero, M.M.; Numa, C.; Bo, M.; Orejas, C.; Garrabou, J.; Cerrano, C.; Kružić, P.; Antoniadou, C.; Aguilar, R.; Kipson, S.; et al. Overview of the Conservation Status of Mediterranean Anthozoans; IUCN: Málaga, Spain, 2017; pp. 1–73. [Google Scholar]
- Bertrand, J.A.; Gil De Sola, L.; Papaconstantinou, C.; Relini, G.; Souplet, A. The general specifications of the MEDITS surveys. Sci. Mar. 2002, 66, 9–17. [Google Scholar] [CrossRef]
- Fiorentini, L.; Dremière, P.Y.; Leonori, I.; Sala, A.; Palombo, V. Efficiency of the bottom trawl used for the Mediterranean International Trawl Survey (MEDITS). Aquat. Living Resour. 1999, 12, 187–205. [Google Scholar] [CrossRef]
- Kenchington, E.; Murillo, F.J.; Cogswell, A.; Lirette, C. Development of Encounter Protocols and Assessment of Significant Adverse Impact by Bottom Trawling for Sponge Grounds and Sea Pen Fields in the NAFO Regulatory Area; NAFO Scientifc Council Report 11/75 Serial No N6005; Agris: Dartmouth, NS, Canada, 2011; Volume 53, pp. 1–51. [Google Scholar]
- Troffe, P.M.; Levings, C.D.; Piercey, G.E.; Keong, V. Fishing gear effects and ecology of the sea whip (Halipteris willemoesi (Cnidaria: Octocorallia: Pennatulacea)) in British Columbia, Canada: Preliminary observations. Aquat. Conserv. 2005, 15, 523–533. [Google Scholar] [CrossRef]
- Ambroso, S.; Gori, A.; Dominguez-Carrió, C.; Gili, J.M.; Berganzo, E.; Teixidó, N.; Greenacre, M.; Rossi, S. Spatial distribution patterns of the soft corals Alcyonium acaule and Alcyonium palmatum in coastal bottoms (Cap de Creus, northwestern Mediterranean Sea). Mar. Biol. 2014, 160, 3059–3070. [Google Scholar] [CrossRef]
- Mastrototaro, F.; Chimienti, G.; Matarrese, A.; Gambi, M.C.; Giangrande, A. Growth and population dynamics of the non-indigenous species Branchiomma luctuosum Grube (Annelida, Sabellidae) in the Ionian Sea (Mediterranean Sea). Mar. Ecol. 2015, 36, 517–529. [Google Scholar] [CrossRef]
- Fryganiotis, K.; Antoniadou, C.; Chintiroglou, C.; Vafidis, D. Redescription of the Mediterranean endemic sea-pen Crassophyllum thessalonicae (Octocorallia: Pteroeididae). Mar. Biodivers. Rec. 2011, 4, 1–3. [Google Scholar] [CrossRef]
- Mastrototaro, F.; Chimienti, G.; Capezzuto, F.; Carlucci, R.; Williams, G. First record of Protoptilum carpenteri (Cnidaria: Octocorallia: Pennatulacea) in the Mediterranean Sea. Ital. J. Zool. 2015, 82, 61–68. [Google Scholar] [CrossRef]




| Set | ||||
|---|---|---|---|---|
| Trawling | 22.0 | 3.7 | −1.3 | 5.6 |
| ROV | 19.9 | 3.5 | −1.1 | 5.5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chimienti, G.; Di Nisio, A.; Lanzolla, A.M.L.; Andria, G.; Tursi, A.; Mastrototaro, F. Towards Non-Invasive Methods to Assess Population Structure and Biomass in Vulnerable Sea Pen Fields. Sensors 2019, 19, 2255. https://doi.org/10.3390/s19102255
Chimienti G, Di Nisio A, Lanzolla AML, Andria G, Tursi A, Mastrototaro F. Towards Non-Invasive Methods to Assess Population Structure and Biomass in Vulnerable Sea Pen Fields. Sensors. 2019; 19(10):2255. https://doi.org/10.3390/s19102255
Chicago/Turabian StyleChimienti, Giovanni, Attilio Di Nisio, Anna M. L. Lanzolla, Gregorio Andria, Angelo Tursi, and Francesco Mastrototaro. 2019. "Towards Non-Invasive Methods to Assess Population Structure and Biomass in Vulnerable Sea Pen Fields" Sensors 19, no. 10: 2255. https://doi.org/10.3390/s19102255





