1. Introduction
Olive growing is a high relevance agricultural activity. With a huge presence in the Mediterranean Basin, where its importance transcends the farming scope to become an actual symbol of its culture and tradition, the olive crop has spread all over the world [
1,
2]. Because of the well-proved health benefits of olive-derived products, and the excellence of its culinary uses, its consumption has considerably risen in recent years. According to IOC (International Olive Council) [
3], table olives consumption has been increased by 173% in the twenty-five years between 1990/91 and 2015/16. Moreover, according to IOC and USDA (United States Department of Agriculture) expectations [
3,
4], olive oil consumption will exceed 3,000,000 tons in 2017/18.
To meet such demand, the olive industry must face multiple challenges. Despite the numbers of its market, olive farming and processing are still mainly performed in a traditional way. Even in Spain, the world largest producer, olive farming is still strongly linked to traditional production systems and low-density olive groves [
5]. This model represents a problem in terms of productivity and profitability. In recent years, super-high-density olive groves, along with increased mechanization, have been introduced as response. Although some indicators suggest that these solutions, based on intensification, can provide the key for economic survival, accurate knowledge about its impact and viability is still yet to be obtained [
6]. Be that as it may, within this context, the enhancement and modernization of the processes, and the introduction of innovative solutions at all levels, are fundamental tasks to be accomplished by this industry.
Fruit sizing is a high-relevance post-harvest task in the food industry [
7]. Sorting fruits and vegetables according to different attributes such as color, mass, size or shape can all be determinants in the quality and pricing of the eventual product. In the olive sector, this is especially relevant [
8,
9]. For table olives, the uniform size, spotless surface or appropriate coloring are determining quality-features as perceived by the final consumer. On the other hand, focusing on olive oil, size and mass of the fruits are used to calculate yield estimations. In any case, the measurement of these parameters is a necessity. However, measuring the whole harvested batch by hand is not an option due to the huge workload involved. Thus, the actual processes to extract this information are based on the study of samples.
In recent years, machine vision techniques have been explored as a valuable tool in food industry. Within the precision agriculture and the horticultural product manufacturing scopes, there is considerable literature regarding the use of image analysis to approach different problems, such as yield estimation, fruit detection, data extraction, sorting and classification or sizing and grading. Thus, Aquino et al. [
10] presented a classification-based algorithm to predict grape yield at early stages from images taken on-the-go directly in the vineyard. Mery et al. [
11] proposed a methodology for the detection, via image segmentation, of different kinds of food previously photographed. Cervantes et al. [
12] developed a comparative analysis of different methods of feature extraction and classification of plant leaves using image processing techniques. Zhang et al. [
13] developed an automatic fruit recognition system based on a split-and-merge algorithm and multiclass support vector machine (SVM). Sa’ad et al. [
14] estimated the mass of mangoes from different photographs of the fruits using thresholding segmentation and provided a classification methodology supported by the information extracted from the results of this segmentation. In the same vein, Mizushima et al. [
15] proposed a method for sorting and grading apples, based on the Otsu’s method and linear SVM. Omid et al. [
16] performed the estimation of volume and mass of citrus fruits through the segmentation of images captured in the laboratory.
Regarding the olive sector, literature focused on the application of machine vision for olive treatment and manufacturing can be consulted. Of special relevance are developments for the classification of fruits according to different characteristics, such as defects on the surface [
17] or the variety [
18,
19], and fruit detection for feature estimation [
20].
This paper proposes an efficient methodology to estimate the maximum/minimum (polar/equatorial) diameter length and mass of olive fruits by means of image analysis. To this end, as a first step, the contrast between the olives and the background is maximized in the images by employing specialized morphological processing. Then, the olives are segmented by automated thresholding based on statistical bimodal analysis. Finally, estimation models for the targeted features are obtained by correlating measurements taken from the segmentations to actual values measured in the laboratory.
The manuscript is structured as follows: Throughout three subsections,
Section 2 describes the experimental design and the data acquisition process, the developed image analysis algorithm and model training for olive characterization. The next section presents the methodology proposed for result evaluation and discusses the achieved results; they have been placed together in order to provide the best understanding of the paper’s research. Finally, the manuscript ends with the main conclusions on the carried-out research.
2. Materials and Methods
2.1. Reference Data and Image Acquisition
Olive fruits from two different varieties were considered for this study: Arbequina and Picual. Samples of both varieties were manually collected in January 2018, in high-density olive orchards located in Lagar Oliveira da Serra (Ferreria do Alentejo, Portugal).
Two populations (one per variety) of 200 olive fruits were selected from the samples previously acquired. Then, they were separated into different groups. Hence, for the Arbequina variety, the following sets were established: A1 (40 fruits), A2 (40 fruits), A3 (40 fruits), A4 (50 fruits) and A5 (30 fruits). For the Picual variety, four groups of 50 olives each were set up and were named as follows: P1, P2, P3 and P4.
Every described set was photographed in the laboratory, spatially distributing olives over a white plastic mat. This durable and deformable material was chosen in an attempt to approximate the type that would be used in a real conveyor belt. For capturing, the LUMIX DMC-GH4 digital single-lens mirrorless camera, equipped with a NMOS sensor, was used (Panasonic, Kadoma, Osaka, Japan). It was set up in manual mode, with an aperture of f/8, an exposure time of 1/500 s, an ISO value of 400 and a focal length of 14 mm. To reproduce an environment close to an actual industrial system, an artificial lightning setup composed of two 500 W halogen floodlights, with a light appearance of 3300 k, was employed for scene illumination. The camera was perpendicularly located above the scene; the lights were placed at the same plane and oriented to the point the camera was focused on.
Figure 1 shows an example of the captured images, which were acquired and saved in JPG format, with
pixels in resolution, a pixel density of 180 ppi and a color depth of 24 bits.
To evaluate the error produced by the estimation models, objective measurements of the major and minor axis length (in millimetres—mm), and mass (in grams—g), were taken for every photographed olive by using:
a KERN PCB 3500-2 precision balance (KERN & Sohn GmbH, Balingen, Germany).
a 0.01 mm-resolution 0.02 mm-accuracy Electronic Digital Vernier Caliper.
The values were annotated and associated to the position of the corresponding olive fruit in the image in which it appeared.
2.2. Image Analysis and Segmentation
The proposed methodology is aimed at automatically extracting from the images features descriptive of the mass and size of the olive fruits. To accomplish this task, the developed algorithm uses techniques based on mathematical morphology and segmentation by clustering-based image thresholding. This algorithm was implemented using MATLAB and Image Processing Toolbox Released 2016a (The MathWorks, Inc., Natick, MA, USA).
2.2.1. Preprocessing
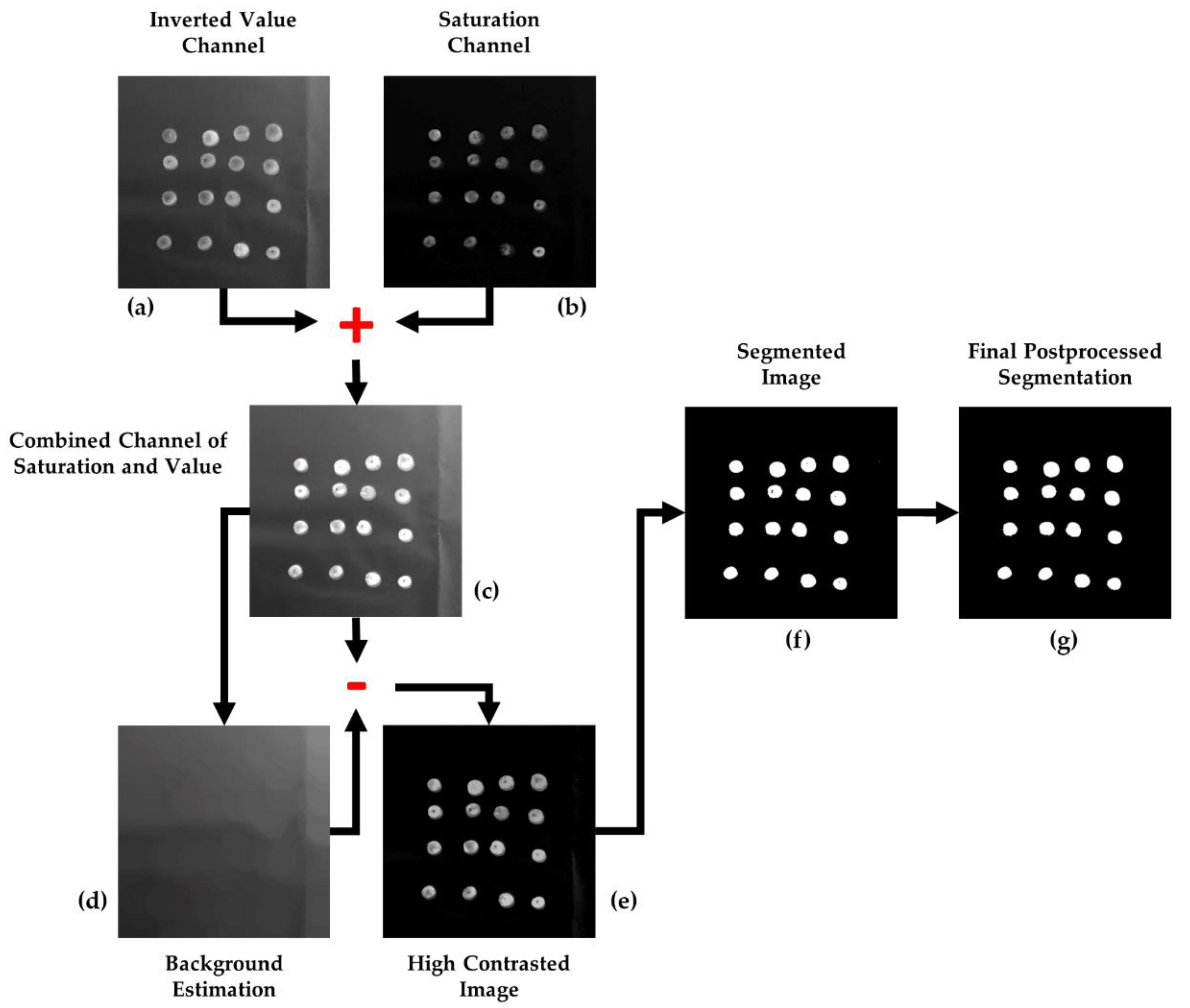
Firstly, images are down-scaled to 40% of its original size using bicubic interpolation for the decreasing of the computational workload. Next, a salt-and-pepper noise reduction is accomplished by applying a gaussian filter (rotationally symmetric gaussian low-pass filter) with a standard deviation of 0.8, and a kernel size of .
Secondly, images are transformed from the native RGB color space to HSV [
21]. After studying the characteristics of the images, it was concluded that the RGB space did not offer an optimal data representation for the purposes of this study. In terms of color, an absence of homogeneity between the olive fruits was detected (especially for the Arbequina variety), which prevented it from being exploited as a distinctive feature. Conversely, the difference between the fruits and the white background in terms of lightness/brightness is remarkable. The level of lightness/darkness of the color of a pixel can be accessed by transforming its RGB values in accordance with a different representation of this color model. Notwithstanding this, basing the process exclusively on light intensity could not yield good segmentation results. Indeed, there were background pixels with lightness values similar to those of olives due to the shadows cast by these fruits. At this point, it was observed that color saturation also provided object differentiation while keeping similar values for background pixels, including both the ones which belonged to a shadow and the ones that did not. Nevertheless, despite this being a partial solution to the shadow problem, the segmentation based merely on saturation couldn’t yield reliable results, leading to olive pixels with saturation levels close to the background values which lacked accuracy. Therefore, neither color saturation nor intensity were found to be fully effective for image segmentation by themselves; however, an accurate combination and processing of both appeared more effective. Due to these reasons, HSV color space provided a solution, as it provides the saturation and value (level of lightness/darkness of the color) information separated into different layers (
S and
V channels, respectively). It is important to note that other existing color spaces are potentially valid according to this scenario, such as HSL [
21] or CIELAB [
22], among others.
2.2.2. Image Segmentation
Once the image is transformed into the HSV color space, the value and saturation channels are isolated into different matrices,
V and
S, respectively. These matrices are transformed and combined into a unique component that it is treated as a grayscale image, which is the one to be segmented. According to this, as a first step, the elements of the
V component are inverted with regard to the maximum possible grey-value, i.e., 255 (for 8-bit per channel image quantification). As such, given
V is the image defined in the interval [0, 255], the image
VINV is the one resulting from the next operation, as can be examined in
Figure 2a:
Considering the
V channel as a greyscale image, the aim of this transformation is to set the higher grey values to olive pixels and, consequently, the lower values to the background, which becomes the darkest part of the image. Then, the saturation layer (
Figure 2b) is combined to the outcome of this transformation, as is shown in
Figure 2c, looking forward to improving the contrast between the background and foreground and to complement information from both sources:
Next, with the purpose of obtaining a background estimation, a morphological opening is applied to
:
where
β is a 50-pixel-radius disk-shaped structuring element, large enough to contain any olive, and
δ and
ε are the basic morphological operations of dilation and erosion, respectively [
23]. The result of this operation can be checked in
Figure 2d. Then, the values of the background estimation are subtracted from
, thus computing a high-contrast image:
The outcome of this operation,
, is the grayscale image to be segmented by binarization. To automatically set an optimum global threshold, the clustering-based method proposed by
Otsu [
24] was selected. This method starts from the premise that the image contains two normal-like distributions of pixels, corresponding to the foreground and the background. Then, the threshold is decided as that which maximizes the inter-class- or minimizes the intra-class-variance to optimize separation. This approach explodes the characteristics of image
, which is the result of an image processing aimed at strengthening contrast between the olives and the background, and at homogenizing the latter to favor binarization using a global threshold. Therefore, by applying the Otsu’s method to
, the threshold
thresh is obtained and applied to undertake its binarization as:
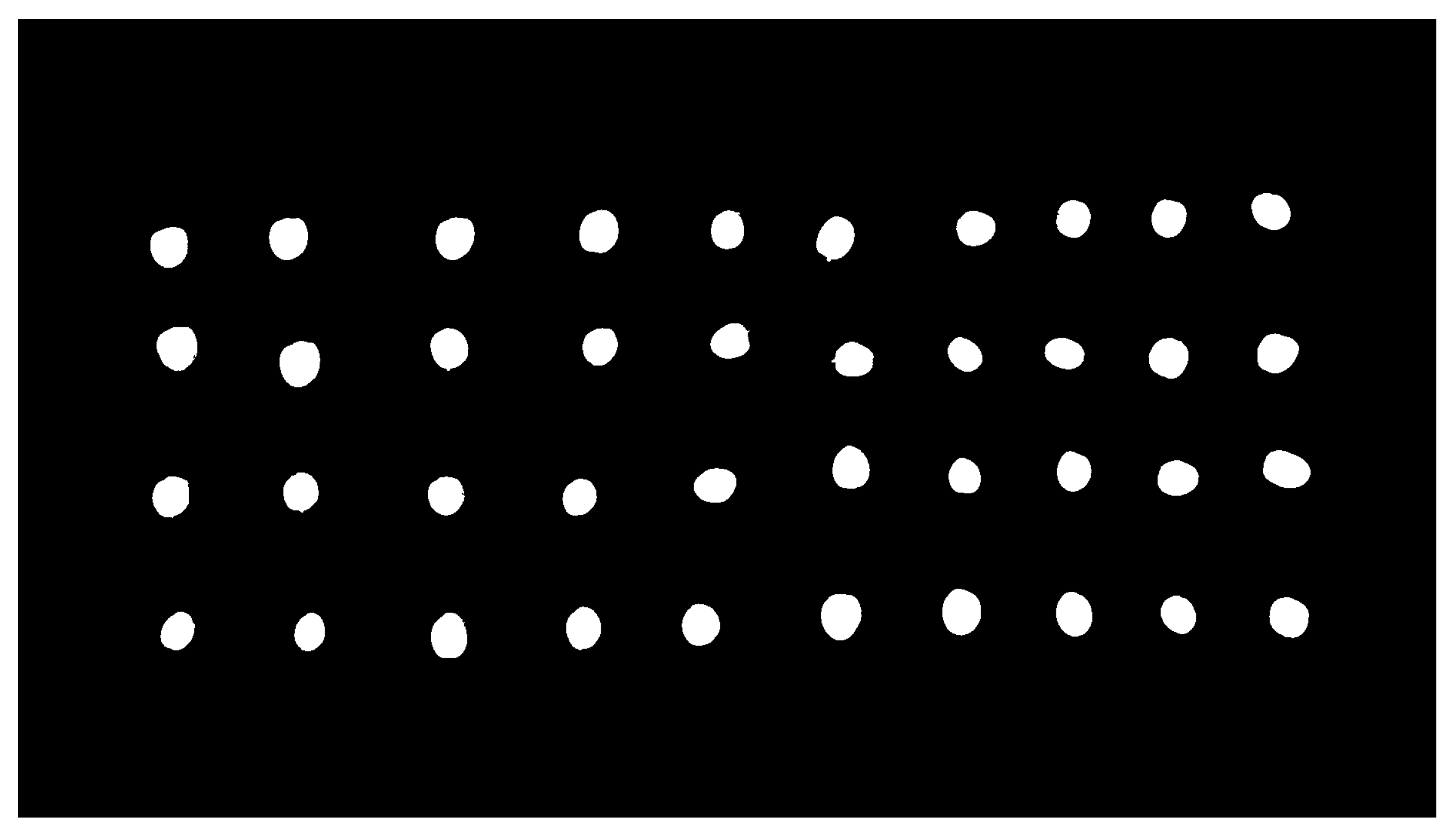
The result of the described methodology for olive fruit segmentation can be analyzed in
Figure 2 and
Figure 3.
2.2.3. Postprocessing
As a last step, some morphological transformations are appealed to improve the final segmentation result. First, false positives filtering is addressed by eliminating those connected components that are too abnormally small to be considered as olive fruits. Mathematically:
where
γ is the morphological opening with a disk-shaped structuring element
β with a radius of 3 pixels.
Finally, a flood-fill operation is applied to eliminate false negatives represented by the small holes which have
emerged inside some fruit-corresponding connected components (the holes derive from points of maximum reflection of light, because of the convex surface of the fruits).
where
R is the morphological reconstruction operation, which consists on the iterative erosion (
ε) of the image
regarding to
, using a unitary structuring element, until idempotence:
where
The corrective effect of this postprocessing is shown in
Figure 2g.
2.3. Estimation Model Training
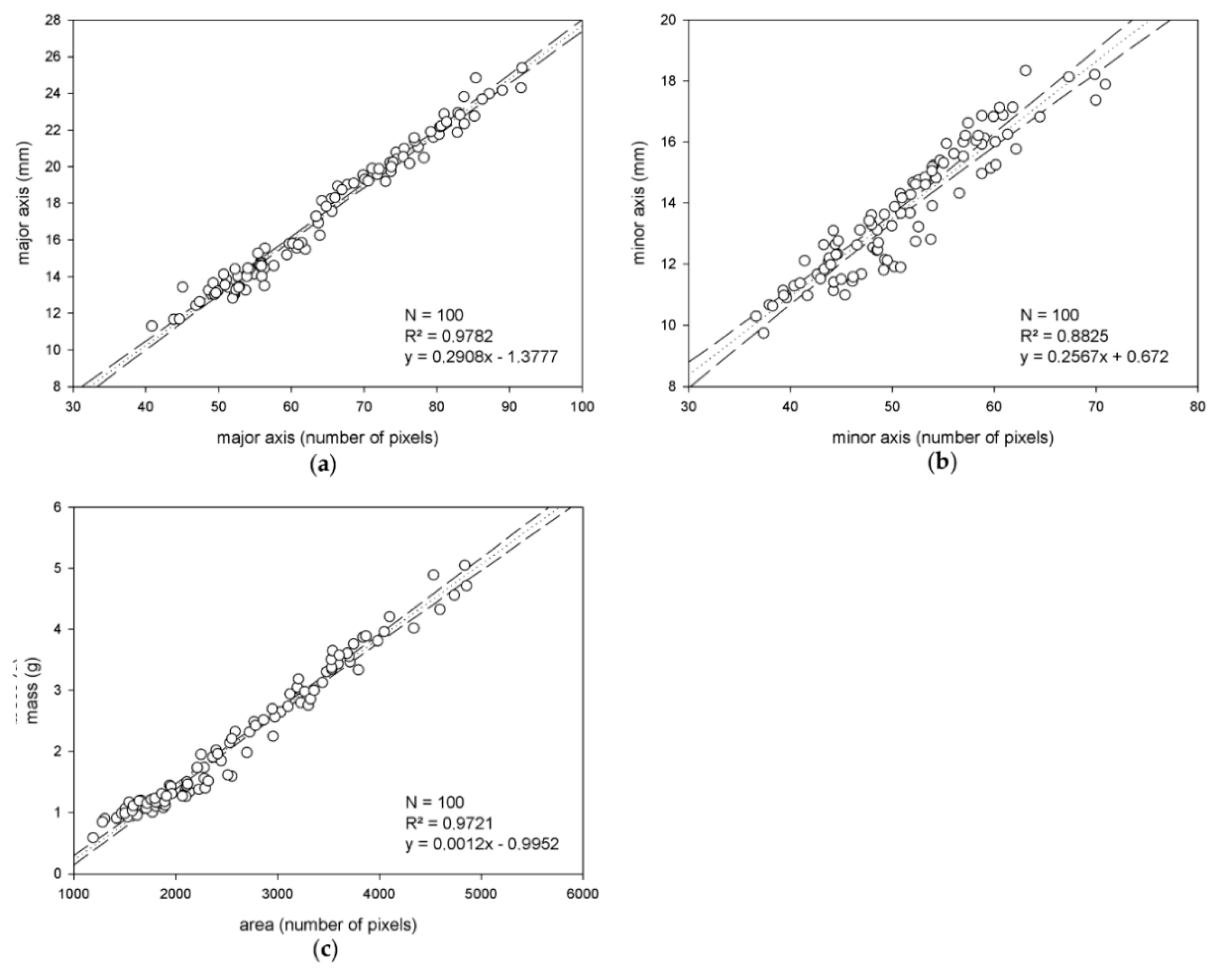
The goal here is to extract descriptive data from the segmented images to build estimation models for olive major and minor axis length, and mass. To this end, the binarized images allow us to work with the connected components representing the different olive fruits. First, to characterize the minor and major axis of the olives, for every component, the ellipse has the same normalized second central moments as it is being computed. Using this method, the major and minor olive axis are approximated to the major and minor axis of this ellipse, respectively, and their length in pixels is used for size estimation. On the other hand, the area of the segmented connected components, calculated as the number of constituent pixels (using 8-connectivity), is selected as a feature to estimate olive mass.
Once this information is extracted, for each of the two considered varieties, a population of 50 individuals/olives is selected as the training set; the remaining 150 individuals are kept for external validation. These training sets are representative of the variability of the samples regarding the features under study. Next, the measurements of the major and minor axis length, and mass, corresponding to these populations and extracted automatically as specified above from the segmented images, are compared to the objective measurements taken in the laboratory. Thus, via regression analysis, linear estimation models for the targeted magnitudes and specific to each variety are yielded. Additionally, variety independent models for the magnitudes are also calculated by joining the two training sets from the two varieties and applying the same described procedure.
4. Conclusions
In the present paper, a method based on image analysis techniques has been developed for estimating the size and mass of olive fruits. The results underscore the robustness and accuracy of the algorithm this method is based on. Moreover, they support its viability for the development of sorting and grading systems for the olive industry.
In accordance with the results, the segmentation algorithm showed a noticeably good performance in the image segmentation binarization task when compared to ground-truth images. Additionally, it was able to detect the exact number of fruits that appeared in every treated image, thus highlighting the accuracy of the process. It is also interesting to note the steadiness of the method dealing with two different olive fruit varieties, as this increases confidence in its applicability to other cultivars. Nevertheless, future trials will focus on analyzing this aspect of the method to verify this generality. Also, these trials could explore different lightning systems, such as diffuse illumination, which could improve the image acquisition task by minimizing the shadows cast by the fruits, thus enabling more reliable segmentation results.
Regarding the estimation of the major and minor axis, and mass of olives, accurate results were measured, which do not indicate the necessity of exploring non-linear modeling to this effect. Especially remarkable is the analyzed behavior of the variety-independent models, which showed comparable, or even better, performance than specific models. This outcome supports their use in the pursuit of applicability and generalization. Notwithstanding this, future investigations will pursue the verification of this conclusion with studies that include samples from more varieties. Moreover, further and wider investigations will also be conducted to more confidently quantify the impact of pixel weighing linearization for mass estimation.
On the other hand, there is a requirement for the proposed methodology to be applied, in terms of the disposition with the olive fruits that are placed on the images. Thus, it is necessary that a certain minimum distance be maintained between every pair of fruits. This fact does not imply a problem in a real scenario, where a non-flat belt conveyor equipped with cleavages can be used, which provides a way to keep the fruits separated from each other. Nevertheless, further work might explore the enhancement of the image-binarization method presented, with the purpose of making possible a reliable segmentation that will correctly work in a scenario in which olives appear to be touching each other. Notwithstanding, it would probably require a considerable increase in algorithm complexity, so it remains to be determined if it could satisfy the working conditions of a real-time system.
The presented solution comprises a promising starting point to develop sorting and grading technologies based on image analysis, which would provide high value for the olive-manufacturing industry.