Metal Nanoparticles/Porous Silicon Microcavity Enhanced Surface Plasmon Resonance Fluorescence for the Detection of DNA
Abstract
:1. Introduction
2. Experimental Details
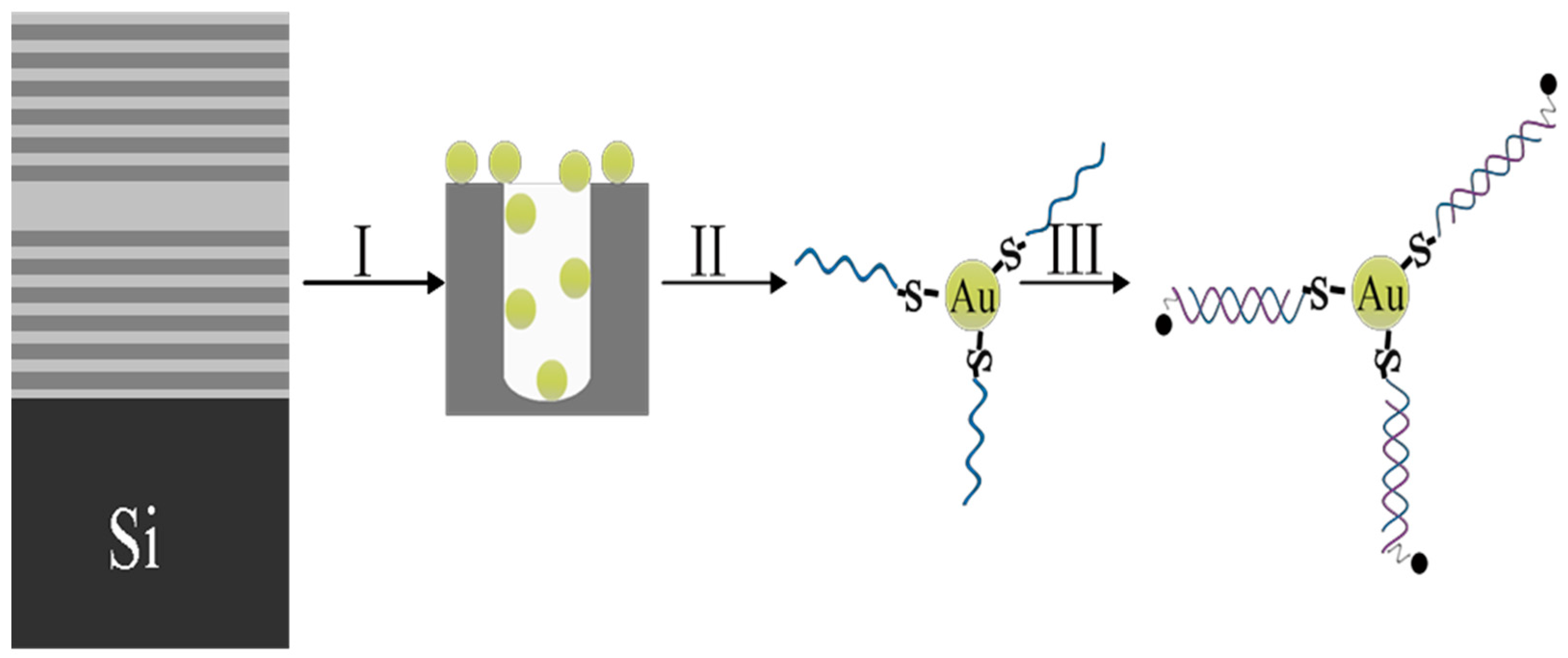
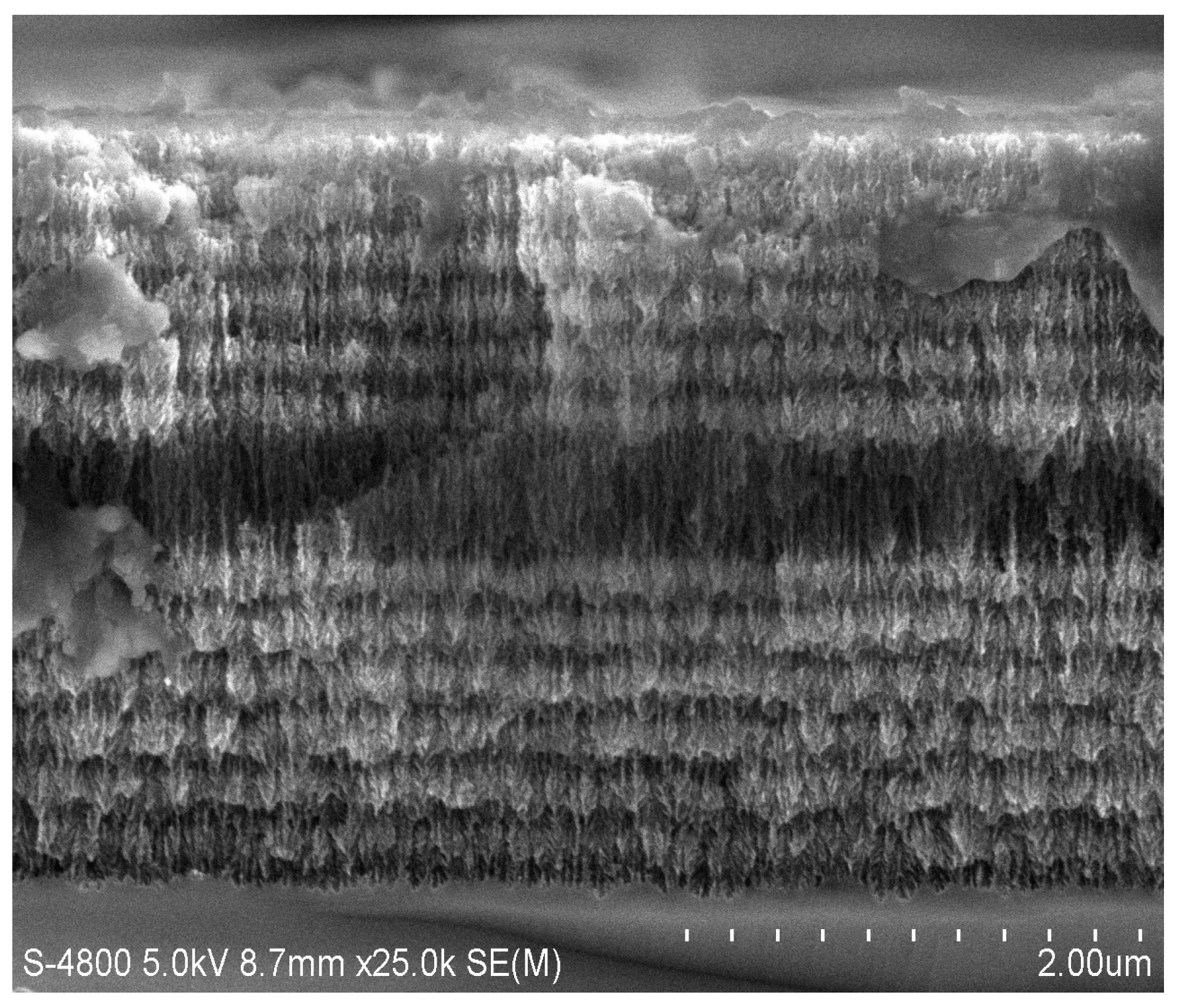
2.1. Fabrication of PSiMC
2.2. The Functionalization of PSi
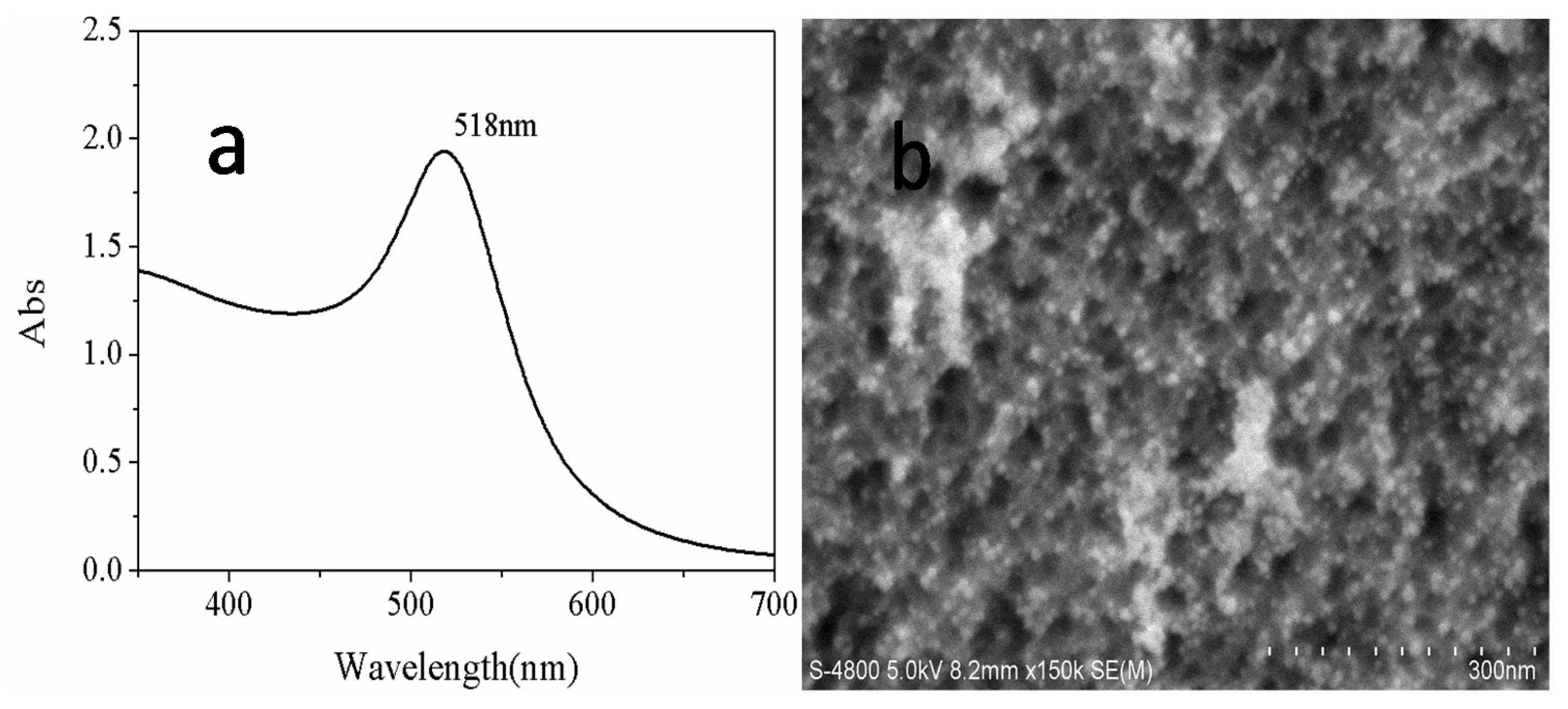
2.3. The Fabrication of Gold Nanoparticles
2.4. DNA Fragment linked to Metal NPs
2.5. Measurements
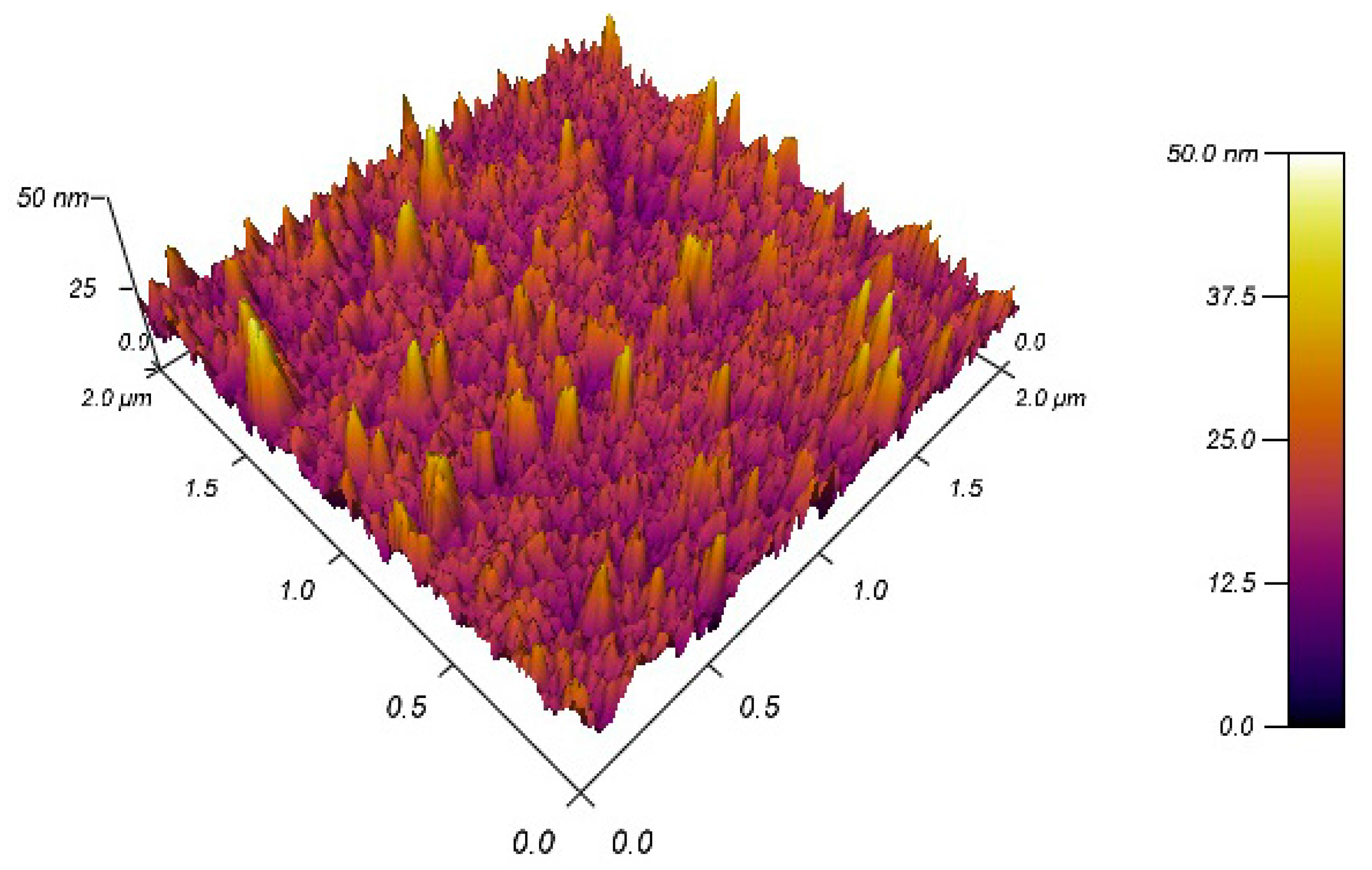
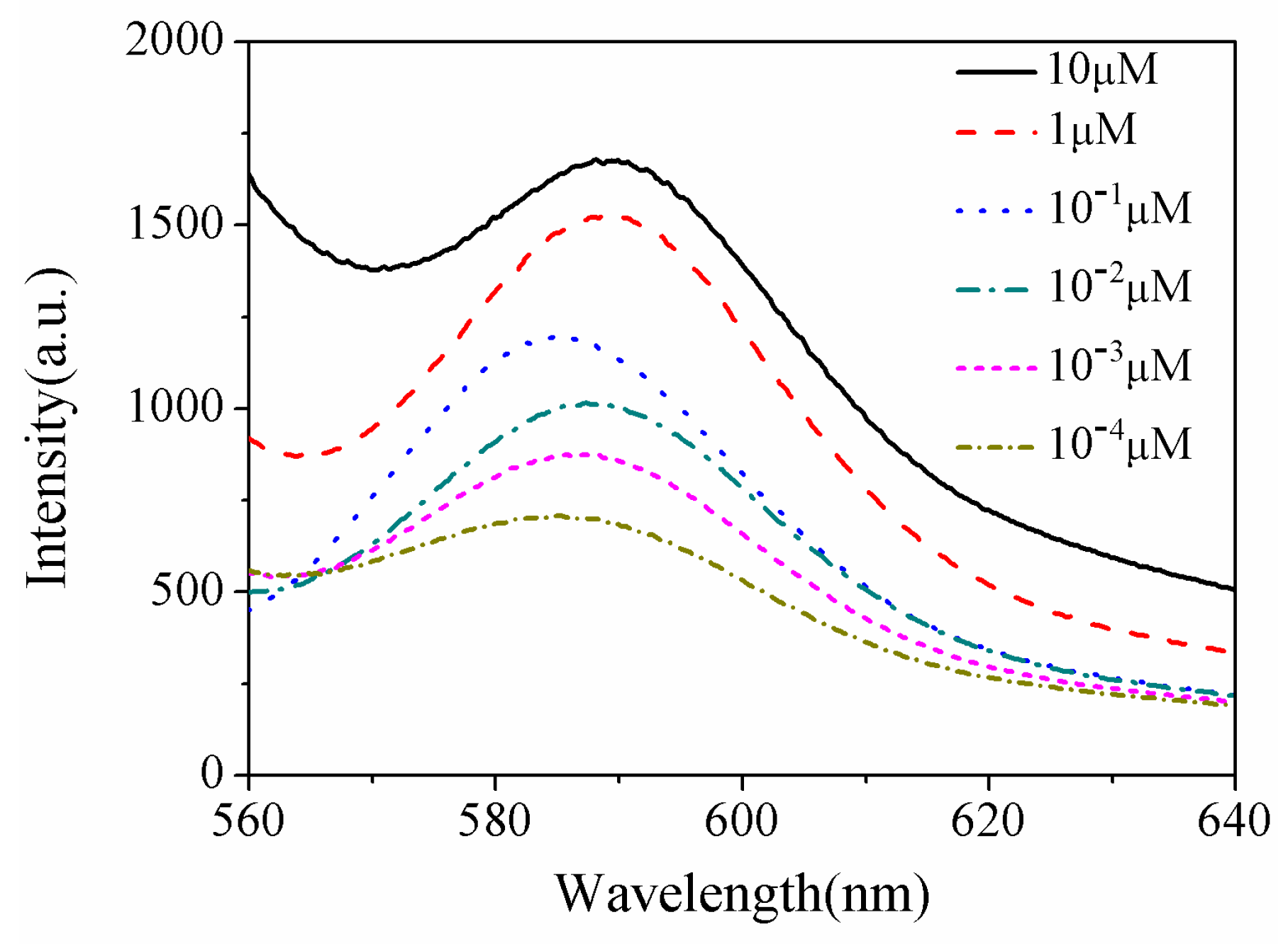
3. Results and Discussion
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Haidary, S.M.; Mohammed, A.B.; Córcoles, E.P.; Ali, N.K.; Ahmad, M.R. Effect of coatings and surface modification on porous silicon nanoparticles for delivery of the anticancer drug tamoxifen. Microelectron. Eng. 2016, 161, 1–6. [Google Scholar] [CrossRef]
- Hiraoui, M.; Haji, L.; Guendouz, M.; Lorrain, N.; Moadhen, A.; Oueslati, M. Towards a biosensor based on anti resonant reflecting optical waveguide fabricated from porous silicon. Biosens. Bioelectron. 2012, 36, 212–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guinan, T.M.; Abdelmaksoud, H.; Voelcker, N.H. Rapid detection of nicotine from breath using desorption ionisation on porous silicon. Chem. Commun. 2017, 53, 5224–5226. [Google Scholar] [CrossRef] [PubMed]
- Ensafi, A.A.; Abarghoui, M.M.; Rezaei, B. Metal (Ni and Bi) coated porous silicon nanostructure, high-performance anode materials for lithium ion batteries with high capacity and stability. J. Alloys Compd. 2017, 712, 233–240. [Google Scholar] [CrossRef]
- Gaur, G.; Koktysh, D.S.; Weiss, S.M. Immobilization of Quantum Dots in Nanostructured Porous Silicon Films: Characterizations and Signal Amplification for Dual-Mode Optical Biosensing. Adv. Func. Mater. 2013, 23, 3604–3614. [Google Scholar] [CrossRef]
- Jane, A.; Dronov, R.V.; Hodges, A.; Voelcker, N.H. Porous Silicon Biosensors on the Advance. Trends Biotechnol. 2009, 27, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Pacholski, C. Photonic Crystal Sensors Based on Porous Silicon. Sensors 2013, 13, 4694–4713. [Google Scholar] [CrossRef] [PubMed]
- Sailor, M.J.; Wu, E.C. Photoluminescence-based sensing with porous silicon films, microparticles, and nanoparticles. Adv. Func. Mater. 2010, 19, 3195–3208. [Google Scholar] [CrossRef]
- Jeffrey, M.L.; Grace, M.C.; Julie, L.H.; Michael, J.S. Reversible luminescence quenching of porous Si by solvents. J. Am. Chem. Soc. 1992, 114, 1911–1912. [Google Scholar]
- Lv, C.; Jia, Z.; Lv, J. High Sensitivity Detection of CdSe/ZnS Quantum Dot-Labeled DNA Based on N-type Porous Silicon Microcavities. Sensors 2017, 17, 80. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Liu, C.; He, H. Surface plasmon enhanced photoluminescence from porous silicon nanowires decorated with gold nanoparticles. RSC Adv. 2016, 6, 59395–59399. [Google Scholar] [CrossRef]
- Mikac, L.; Ivanda, M.; Gotić, M.; Maksimović, A.; Trusso, S.; D’Andrea, C. Metal nanoparticles deposited on porous silicon templates as novel substrates for SERS. Croatica Chemica Acta 2015, 88, 1–8. [Google Scholar] [CrossRef]
- Shen, W. Hierarchical optical antenna: Gold nanoparticle-modified photonic crystal for Highly-sensitive label-free DNA detection. J. Mater. Chem. 2012, 22, 8127–8133. [Google Scholar] [CrossRef]
- Park, S. Nanosilver Colloids-Filled Photonic Crystal Arrays for Photoluminescence Enhancement. Nanoscale Res. Lett. 2010, 5, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Jia, Z.; Lv, X.; Lv, C.; Shi, F. Enhancement of QDs’ fluorescence based on porous silicon Bragg reflector. Physica B 2015, 457, 263–268. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Lv, J.; Jia, Z.H. Efficient Fluorescence Resonance Energy Transfer between Quantum Dots and Gold Nanoparticles Based on Porous Silicon Photonic Crystal for DNA Detection. Sensors 2017, 17, 1078. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; An, Z.; Ren, Q.; Wang, H.; Chen, Z.; Shen, X. Localized-surface-plasmon enhanced luminescence from porous silicon by gold nanoparticles. In Proceedings of the 2010 3rd International Nanoelectronics Conference (INEC), Hong Kong, China, 3–8 January 2010. [Google Scholar] [CrossRef]
- Cohen, A.; Bocobza, S.; Veksler, I.; Gabdank, I.; Barash, D.; Aharoni, A. Colored porous silicon as support for plasmonic nanoparticles. J. Appl. Phys. 2012, 111, 21793. [Google Scholar]
- Mulazimoglu, E.; Nogay, G.; Turan, R.; Emrah, U.H. Enhanced localized surface plasmon resonance obtained in two step etched silicon nanowires decorated with silver nanoparticles. Appl. Phys. Lett. 2013, 103, 267–297. [Google Scholar] [CrossRef]
- Tinsley-Bown, A.M.; Canham, L.T.; Hollings, M.; Anderson, M.H.; Reeves, C.L.; Cox, T.I.; Nicklin, S.; Squirrell, D.J.; Perkins, E.; Hutchinson, A.; et al. Tuning the Pore Size and Surface Chemistry of Porous Silicon for Immunoassays. Phys. Status Solidi A 2000, 182, 547–553. [Google Scholar] [CrossRef]
- Sciacca, B.; Secret, E.; Pace, S.; Gonzalez, P.; Geobaldo, F.; Quignard, F.; Cunin, F. Chitosan-Functionalized Porous Silicon Optical Transducer for the Detection of Carboxylic Acid-Containing Drugs in Water. J. Mater. Chem. 2011, 21, 2294–2302. [Google Scholar] [CrossRef]
- Kwon, D.H.; An, H.H.; Kim, H.S. Electrochemical albumin sensing based on silicon nanowires modified by gold nanoparticles. Appl. Surf. Sci. 2011, 257, 4650–4654. [Google Scholar] [CrossRef]
- Williams, S.E.; Davies, P.R.; Bowen, J.L. Controlling the Nanoscale Patterning of AuNPs on Silicon Surfaces. Nanomaterials 2013, 3, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Frens, G. Controlled Nucleation for the Regulation of the Particle Size in Monodisperse Gold Suspensions. Nat. Phys. Sci. 1973, 241, 20–22. [Google Scholar] [CrossRef]
- Estrada, L.C.; Roberti, M.J.; Simoncelli, S. Detection of low quantum yield fluorophores and improved imaging times using metallic nanoparticles. J. Phys. Chem. B 2012, 116, 2306. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Moore, J.; Laulhe, S. Fluorophore-gold nanoparticle complex for sensitive optical biosensing and imaging. Nanotechnology 2012, 23, 095501. [Google Scholar] [CrossRef] [PubMed]
- Novotny, L.; Bharadwaj, P. Spectral dependence of single molecule fluorescence enhancement. Opt. Express 2007, 15, 14266–14274. [Google Scholar]
- Setzu, S.; Létant, S.; Solsona, P.; Romestain, R.; Vial, J.C. Improvement of the luminescence in p-type as-prepared or dye impregnated porous silicon microcavities. J. Lumin. 1998, 80, 129–132. [Google Scholar] [CrossRef]
- Palestino, G.; Agarwal, V.; Aulombard, R.; Pérez, E.; Gergely, C. Biosensing and protein fluorescence enhancement by functionalized porous silicon devices. Langmuir ACS J. Surf. Colloids 2008, 24, 13765–13771. [Google Scholar] [CrossRef] [PubMed]
- Toma, K.; Vala, M.; Adam, P. Compact surface plasmon-enhanced fluorescence biochip. Opt. Express 2013, 21, 10121–10132. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Dong, J.K.; Kim, K. Surface-enhanced plasmon resonance detection of nanoparticle-conjugated DNA hybridization. Appl. Opt. 2010, 49, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Baillargeat, D.; Ho, H.P. Nanomaterials enhanced surface plasmon resonance for biological and chemical sensing applications. Chem. Soc. Rev. 2014, 43, 3426. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Sreekanth, K.V.; Shang, J. Graphene-Gold Metasurface Architectures for Ultrasensitive Plasmonic Biosensing. Adv. Mater. 2015, 27, 6163–6169. [Google Scholar] [CrossRef] [PubMed]
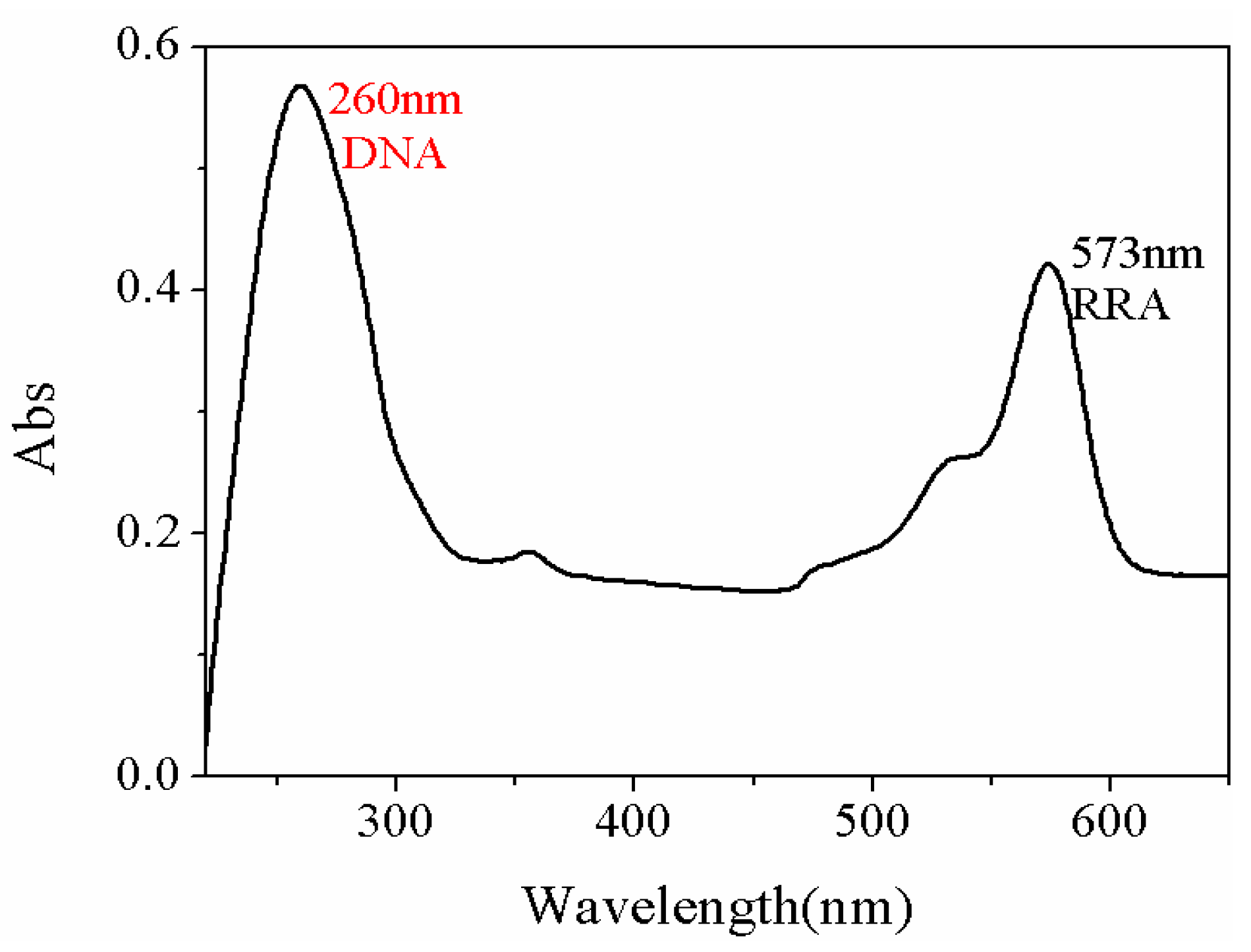

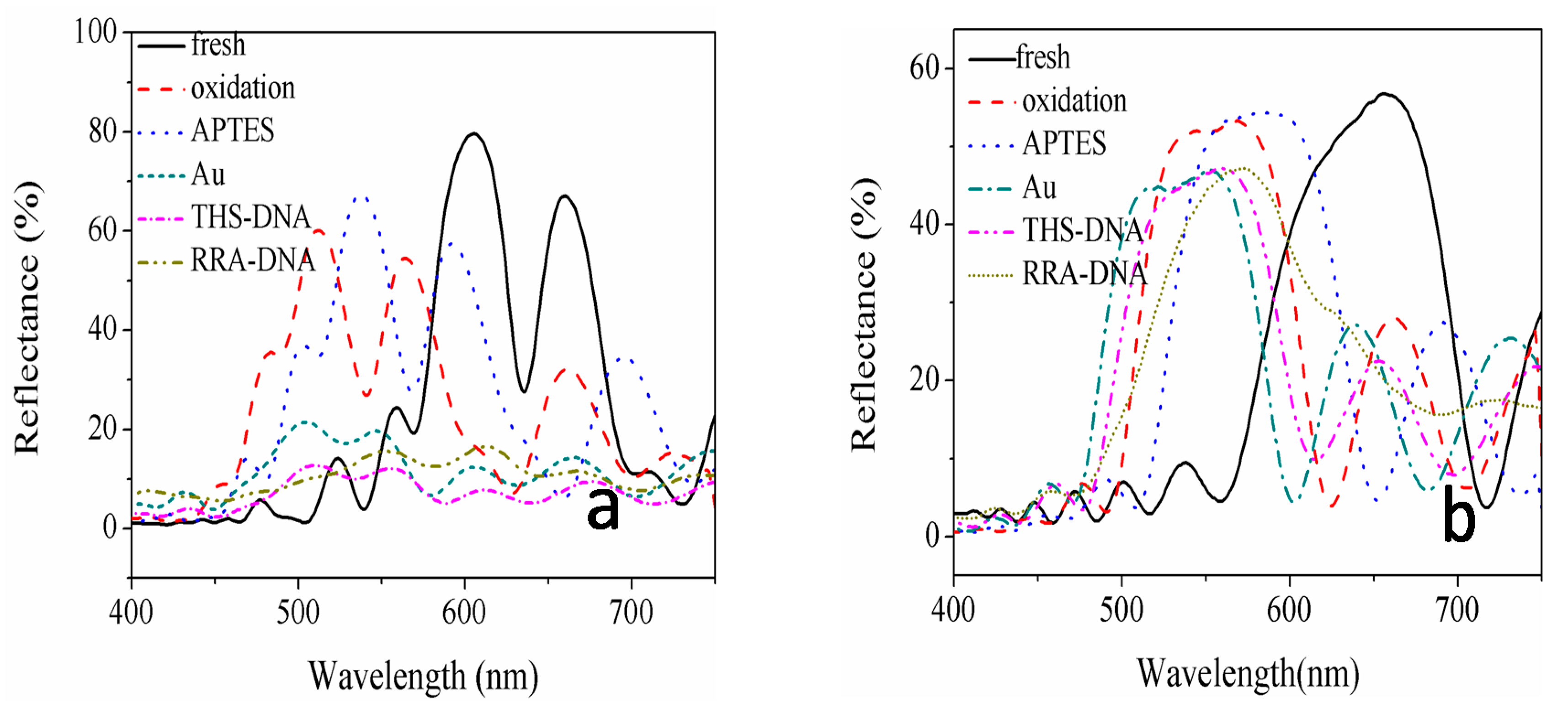

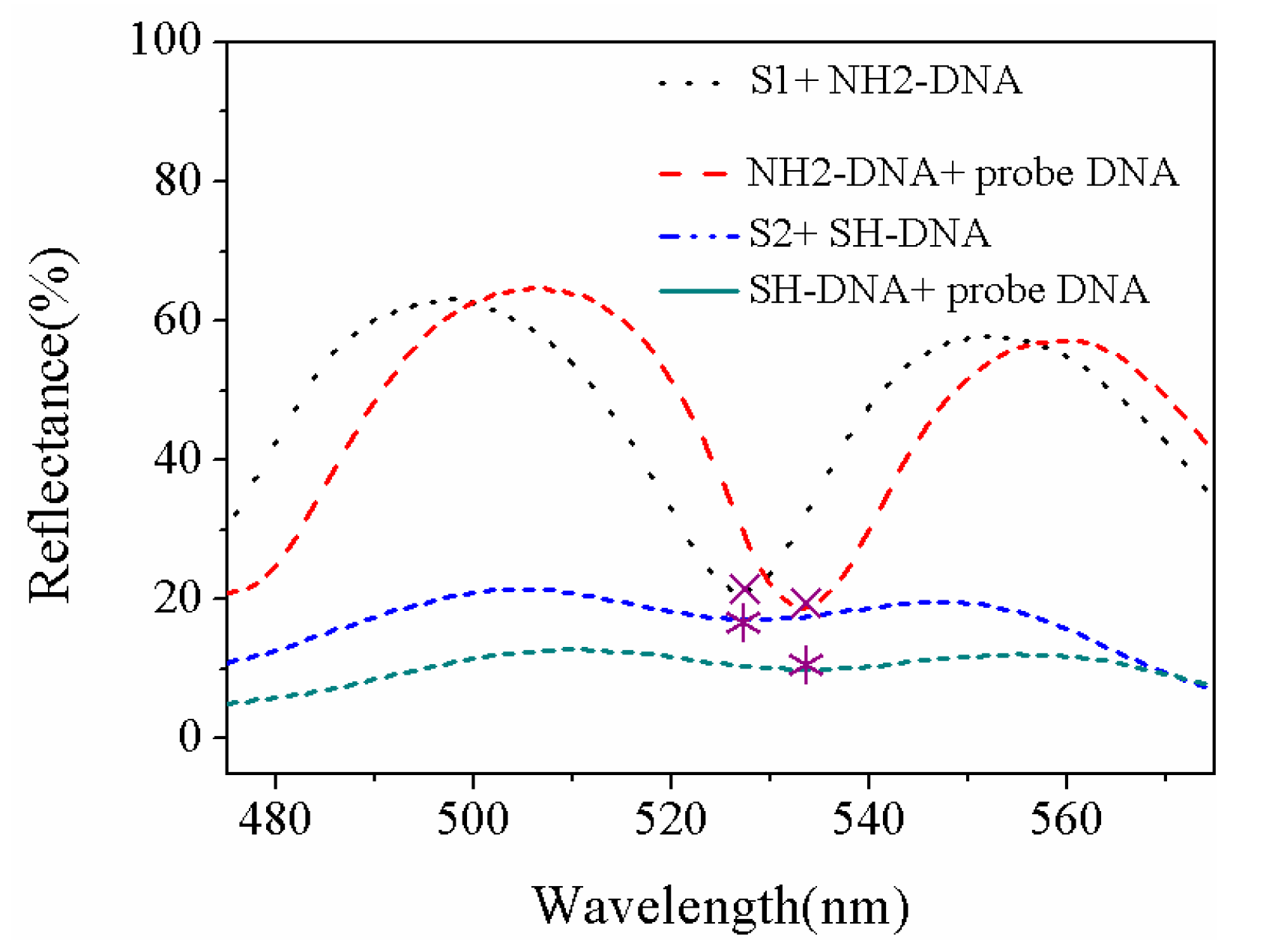
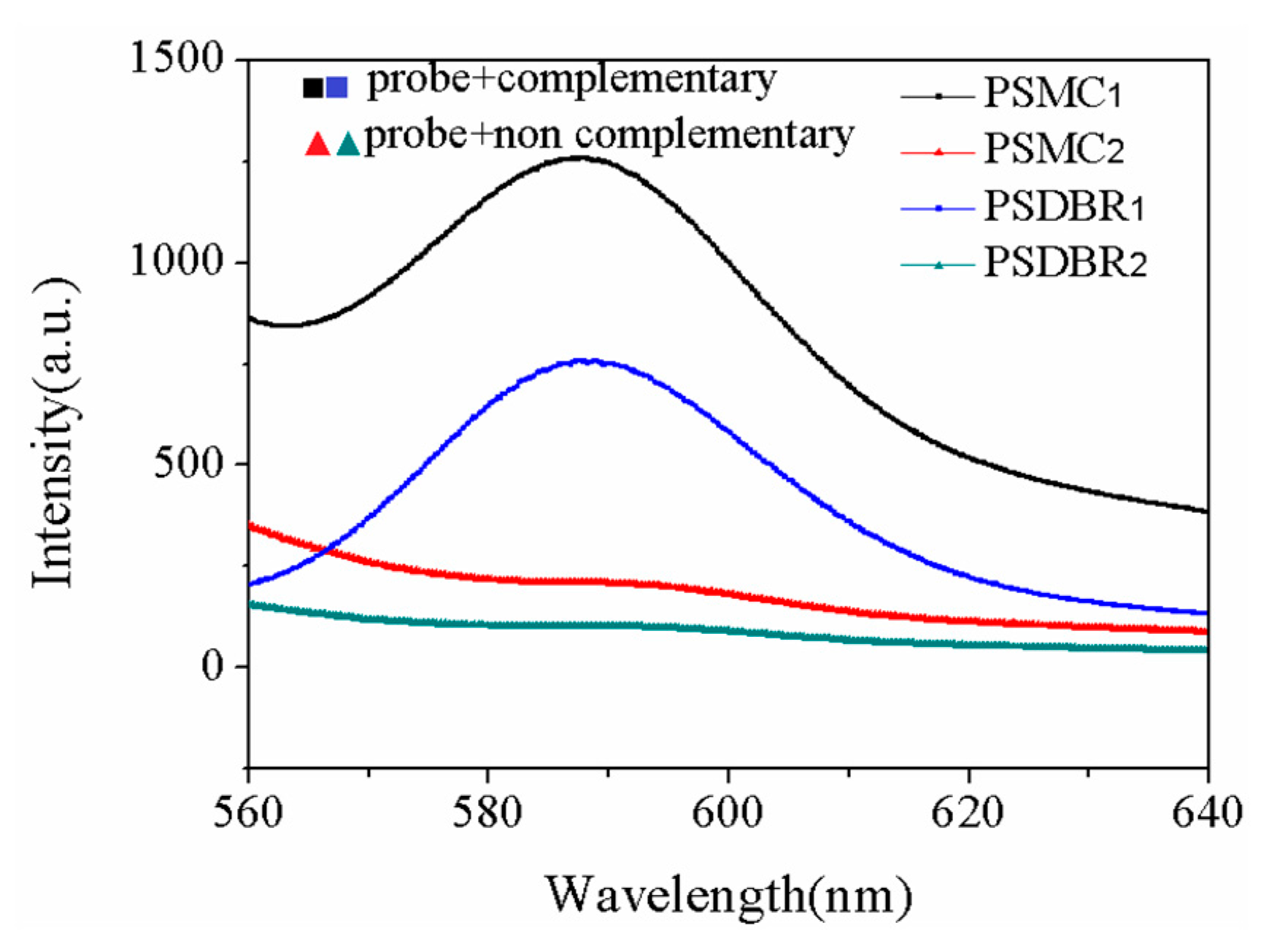

| Sample | Current Density | Corrosion Sequence |
|---|---|---|
| MC | Layer A: 110 mA/cm2 | (AB)6AAAA(BA)6 |
| Layer B: 60 mA/cm2 | ||
| Cavity layer: 110 mA/cm2 | ||
| Bragg reflector | Layer A: 110 mA/cm2 | (AB)14 |
| Layer B: 60 mA/cm2 |
| Sample Number | DNA Fragment |
|---|---|
| Sample 1 | Target DNA fragment: THS-5′-GGCCTATCAGCTTG-3′ |
| Probe1 DNA fragment: RRA-5′-CAAGCTGATAGGCC-3′ | |
| Sample 2 | Target DNA fragment: THS-5′-GGCCTATCAGCTTG-3′ |
| Probe2 DNA fragment: RRA-5′-GGCCTATCAGCTTG-3′ | |
| Sample 3 | Target DNA fragment: NH2-5′-GGCCTATCAGCTTG-3′ |
| Probe3 DNA fragment: RRA-5′-CAAGCTGATAGGCC-3′ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Jia, Z. Metal Nanoparticles/Porous Silicon Microcavity Enhanced Surface Plasmon Resonance Fluorescence for the Detection of DNA. Sensors 2018, 18, 661. https://doi.org/10.3390/s18020661
Wang J, Jia Z. Metal Nanoparticles/Porous Silicon Microcavity Enhanced Surface Plasmon Resonance Fluorescence for the Detection of DNA. Sensors. 2018; 18(2):661. https://doi.org/10.3390/s18020661
Chicago/Turabian StyleWang, Jiajia, and Zhenhong Jia. 2018. "Metal Nanoparticles/Porous Silicon Microcavity Enhanced Surface Plasmon Resonance Fluorescence for the Detection of DNA" Sensors 18, no. 2: 661. https://doi.org/10.3390/s18020661





